In this application note, we demonstrate that a two-dimensional chromatographic assay (2D LC or 2D UPLC) for mulitple reaction monitoring (MRM) quantification in protein bioanalysis can provide up to a three-fold increase in sensitivity compared to one-dimensional LC/MRM. Additionally, high pH/low pH RP/RP separations can significantly reduce analyte suppression.
The 2D LC/MRM assay can provide up to a three-fold increase in sensitivity compared to 1D LC/MRM. High pH/low pH RP/RP 2D LC separations can significantly reduce analyte suppression in protein bioanalysis.
Quantification of therapeutic proteins in serum without analyte pre-fractionation can offer some advantages in terms of reducing the assay costs and simplifying the sample preparation workflow. Analyte isolation (typically performed by immunoaffinity) requires additional purification steps and uses expensive isotopically labeled protein standards to account for analyte recovery.
An alternative approach is the use of an LC system with greater chromatographic resolution, such as multi-dimensional LC. The multiple reaction monitoring (MRM) assays designed for measuring protein therapeutics in complex serum digests produced without analyte fractionation can be enhanced by multi-dimensional chromatography.
In particular, two-dimensional reversed phase/reversed phase (RP/RP) chromatography has been of significant interest in the bioanalysis community in recent years.1-5 The major driving force behind the adoption of 2D-chromatographic methods is demonstrated improvement in the separation of the analyte of interest from other sample components in order to reduce suppression of the analyte signal. 2D-chromatographic techniques are responsible for increased sensitivities compared to one-dimensional LC methods using the same amount of sample.
Trastuzumab (herceptin) is a humanized IgG1 kappa monoclonal antibody (mAb). The antibody was obtained through genetic engineering6,7 by joining the constant regions of the human monoclonal antibody with the complementarity-determining regions (CDRs) of a mouse monoclonal antibody able to bind human epidermal growth factor receptor 2 proteins (HER2) receptors. These HER2 receptors belong to a family of human oncoproteins expressed in approximately 25% of invasive breast cancers. Trastuzumab was approved in 1998 by the U.S. Food and Drug Administration (FDA) for the treatment of HER2-overexpressing breast cancers. Trastuzumab is administered by intravenous infusions in clinical doses to produce saturation of the HER2 receptor. For a conventional 4 mg/kg loading dose, followed by 2 mg/kg weekly doses, the mean maximum concentration of trastuzumab in plasma of 22 patients was approximately 70 μg/mL.8
In this application note, we report the development of a highly sensitive 2D LC/MRM assay for trastuzumab in human serum, employing gradient separation at pH 10.0 in the first RP chromatographic dimension, followed by gradient RP separation at pH 2.5 in the second dimension. We demonstrate that two-dimensional high pH/low pH RP/RP chromatography is able to significantly reduce ion suppression in protein bioanalysis.
A stock solution of trastuzumab (150 kDa monoclonal antibody) was spiked with the internal standard (13C15N-isotopically labeled extended peptide GRFTISADTSK), and digested using trypsin to produce a stock solution containing 5 μM of digested trastuzumab and 10 μM of internal standard peptide (FTISADTSK). In parallel, 400 μL of human serum was dispensed in 10 Eppendorf vials (40-μL serum/vial), and digested using trypsin following the same procedure, to produce 200 μL of human serum digest in each vial. The digestion protocol involved sample denaturation (with 0.05% RapiGest at 80 °C for 10 min), disulfide bond reduction (in the presence of 20 mM dithiothreitol (DTT) for 60 min at 60 °C), cysteine alkylation (10 mM iodoacetamide (IAM) for 30 min at room temperature in the dark), and overnight digestion using porcine trypsin (25:1 (w/w) protein to enzyme ratio). Following digestion, half of the sample (100 μL of peptide digest) was diluted 1:1 with a solution containing 4% H3PO4, then loaded onto an Oasis mixed-mode SPE Elution Plate (10 mg/well of 30 μm MCX particles p/n 186000259). Digests were washed with 500 μL of 2% FA and 500 μL of 5% methanol before being eluted with 2 x 200 μL aliquots of 25% ACN in 1.5% NH4OH (pH 10).
Matrix-free digests were prepared by diluting the 5-μM trastuzumab digest with 20 mM ammonium formate (pH 10) to prepare the following concentrations: 0.1, 1.0, 5.0, and 50.0 nM.
The same trastuzumab concentrations (0.1, 1.0, 5.0, and 50.0 nM) were prepared in 20 mM ammonium formate (pH 10) by diluting the stock trastuzumab digest in the human serum digest (using more than 80% of the serum digest matrix for each dilution).
1D LC/MRM
An ACQUITY UPLC I-Class System equipped with an ACQUITY UPLC BEH300 C18 2.1 x 150 mm, 1.7 μm Column (p/n 180003687) was used. The column temperature was maintained at 35 °C, and the flow rate was 0.3 mL/min. Mobile phases contained 0.1% (v/v) formic acid (FA) in water (A) and 0.1% (v/v) FA in acetonitrile (B). Peptides were eluted with a linear gradient from 0% to 35% B in 10 min.
An ACQUITY UPLC H-Class System with 2D Technology configured in the heart-cutting mode was used for twodimensional chromatography. Peptide separations were performed by RP/RP chromatography using the pH of the mobile phases to change the selectivity of the separation.9,10 A diagram of the 2D-LC system is shown in Figure 1. The first dimension separation was performed on a 1.0 x 50 mm column packed with 2.5-μm XBridge C18 pa particles, kept at 35 °C, and operated at 100 μL/min. Mobile phases contained 20 mM ammonium formate in water, pH 10.0 (Solvent A), and 20 mM ammonium formate in 90% ACN (Solvent B). Peptides were eluted with a linear gradient from 0% to 40% B in 5 min, as shown in Figure 1A. The analyte of interest (FTISADTSK) peptide from trastuzumab), eluting between 6.6 and 6.9 min in the first chromatographic dimension, was transferred to the second chromatographic dimension using a switching valve, as shown in Figure 1B. Shortly after analyte transfer (~ 0.1 min later), the gradient for the second chromatographic separation began, as shown in Figure 1C. This separation was performed on a 2.1 x 50 mm column, packed with BEH300 1.7-μm particles, maintained at 35 °C, and operated at 300 μL/min. Mobile phases for low pH separations contained 0.1% formic acid (FA) in water (Eluent A) and 0.1% FA in ACN (Eluent B). Peptides were eluted with a linear gradient from 0% to 30% B in 5 min (gradient started 7 min after sample injection on the first dimension). The total run time for the entire 2D LC method was 15 min.
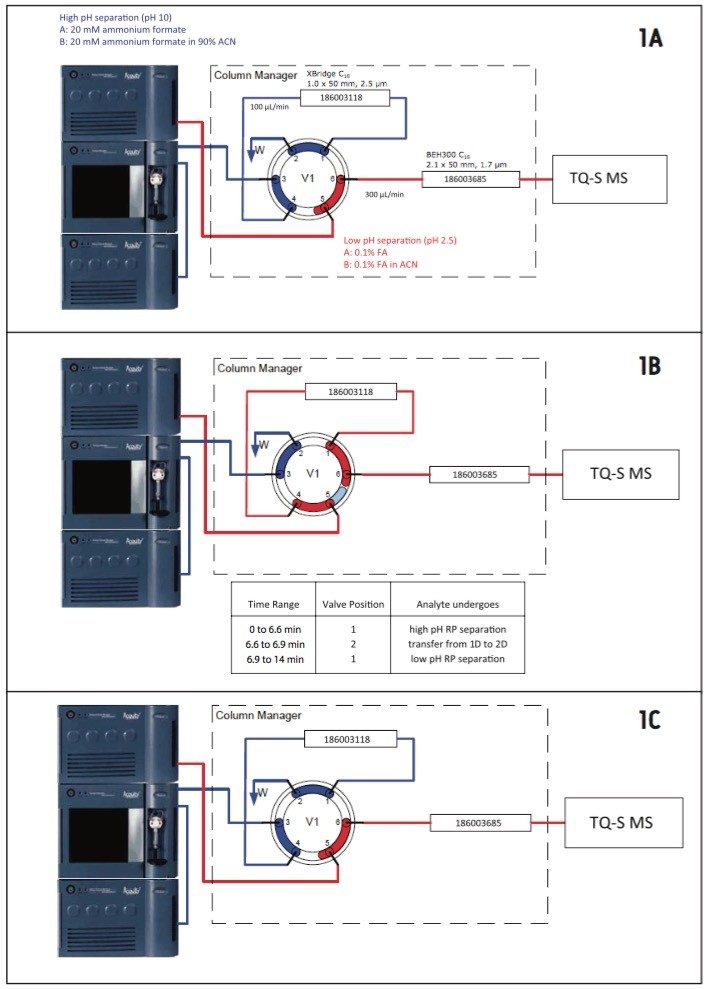
Assays were performed on a Xevo TQ-S Mass Spectrometer operated in MRM positive ion electrospray mode. T he operating parameters were as follows: ESI potential 3.5 kV, CV 28 V, source temperature 120 °C, MS1/MS2 isolation window 0.75 Da (FWHM), 28 eV collision energy. Four MRM transitions were monitored continuously throughout the 2D LC/MRM assay using a dwell time of 100 ms: two MRMs monitored the endogenous signature peptide FTISADTSK from trastuzumab (485.2 → 721.4 for peptide quantification; 485.2 → 608.3 for peptide confirmation), while the other two MRM channels monitored the corresponding 13C15N-isotopically labeled internal standard peptide FTISADTSK (489.2 → 729.4 for peptide quantification; 489.2 → 616.3 for peptide confirmation). In addition to the assays designed for MRM monitoring only, several experiments were performed with RADAR and MRM monitoring. The scans for RADAR monitoring were performed in positive ESI mode over a mass range of m/z = 400 to 1400 with 1 s scans.
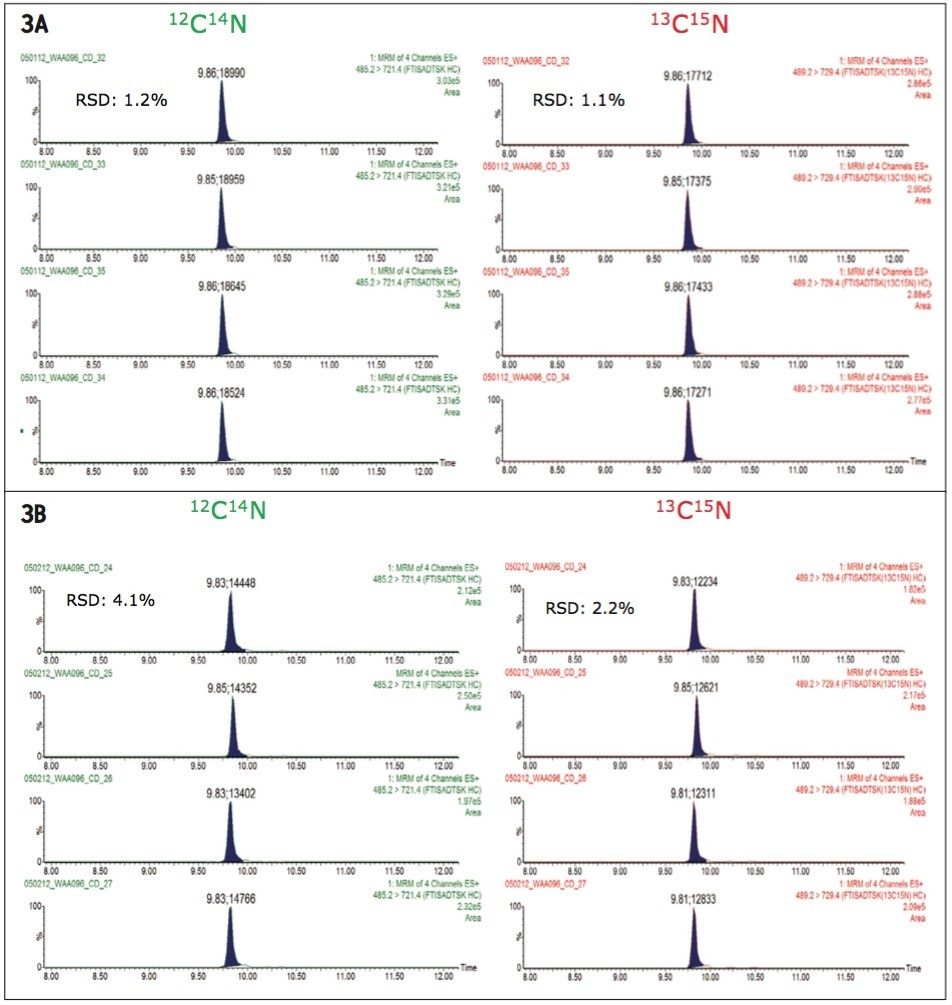
Figure 2 shows the 2D LC/MRM chromatograms of FTISADTSK, the signature peptide from trastuzumab, recorded at four digest concentrations in the range of 0.1 to 50.0 nM. The digest was diluted with 20 mM ammonium formate (pH=10.0) in the absence of serum digest matrix. The assay linearity over the dynamic range investigated (500-fold) is clearly demonstrated by the peak areas displayed in Figure 2. The reproducibility of the 2D assay is shown in Figure 3A where peak areas were recorded for replicate injections (n=4) of the digest containing 5 nM trastuzumab and 10 nN 13C15N-isotopically labeled peptide standard. The overall assay reproducibility was also very good, with the average peak area RSD for all concentrations tested greater than 2%, when four replicate injections were performed for each trastuzumab concentration.
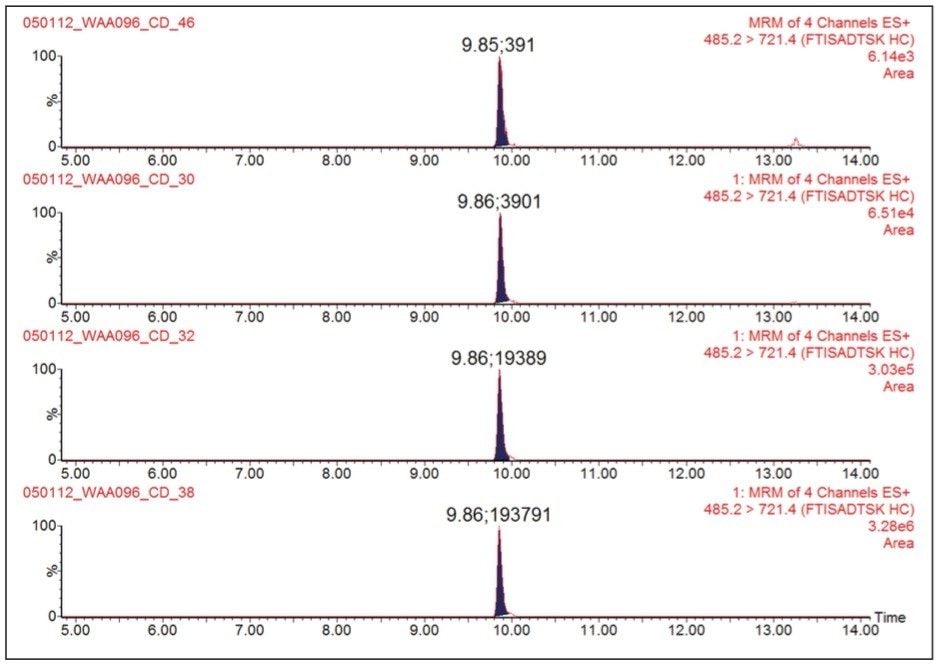
The 2D LC/MRM assay, in the absence of the serum digest matrix, is very sensitive, with the lowest detected trastuzumab concentration determined to be 0.1 nM or 15.0 ng/mL, which is at minimum three orders of magnitude lower than the mean maximum trastuzumab concentration of 70 μg/mL measured in patients’ plasma.8
The sensitivity of the MRM assay was also investigated in the presence of the complex serum digest matrix. Trastuzumab digests were spiked in SPE-cleaned human serum digests and analyzed by 2D LC/MRM, as shown in Figure 3B. This figure reveals that the signals corresponding to the native FTISADTSK peptide, as well as the signals produced by its isotopicallly labeled analogue, were clearly suppressed by co-eluting compounds from serum digest. However, the ratio between the native peptide and the isotopically labeled peptide was not affected by the complex matrix. This is an important observation, as the quantification method is based on comparing the peak area obtained for an unknown trastuzumab concentration to the peak area produced by the IS peptide spiked at a known concentration in the serum sample. Peak area RSDs were greater than 5%, as shown in these chromatograms in Figure 3B. In conclusion, in the 2D LC/MRM experiment, the analyte/IS suppression due to the complex serum digest background was only ~25%.
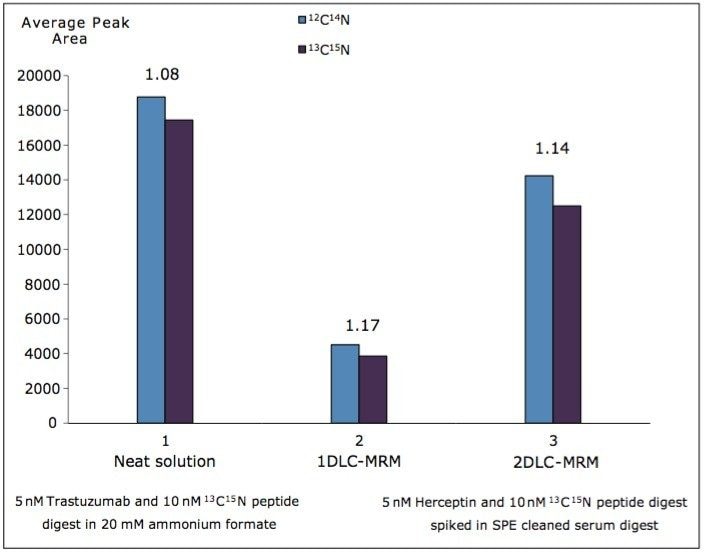
When the trastuzumab digests spiked in SPE-cleaned human serum were analyzed by 1D LC/MRM, the analyte suppression was four-fold higher, as illustrated by the graphs shown Figure 4. The average peak areas of the native and isotopically labeled peptide IS are displayed in this figure for 1D- and 2D-MRM experiments performed for 5 nM trastuzumab digest prepared in neat solvent (20 mM ammonium formate, pH=10.0), as well as in SPE-cleaned human serum. These results clearly indicate that, relative to the quantification of therapeutic proteins in serum, the 2D LC/MRM method can provide up to a three-fold increase in sensitivity compared to conventional 1D LC/MRM.
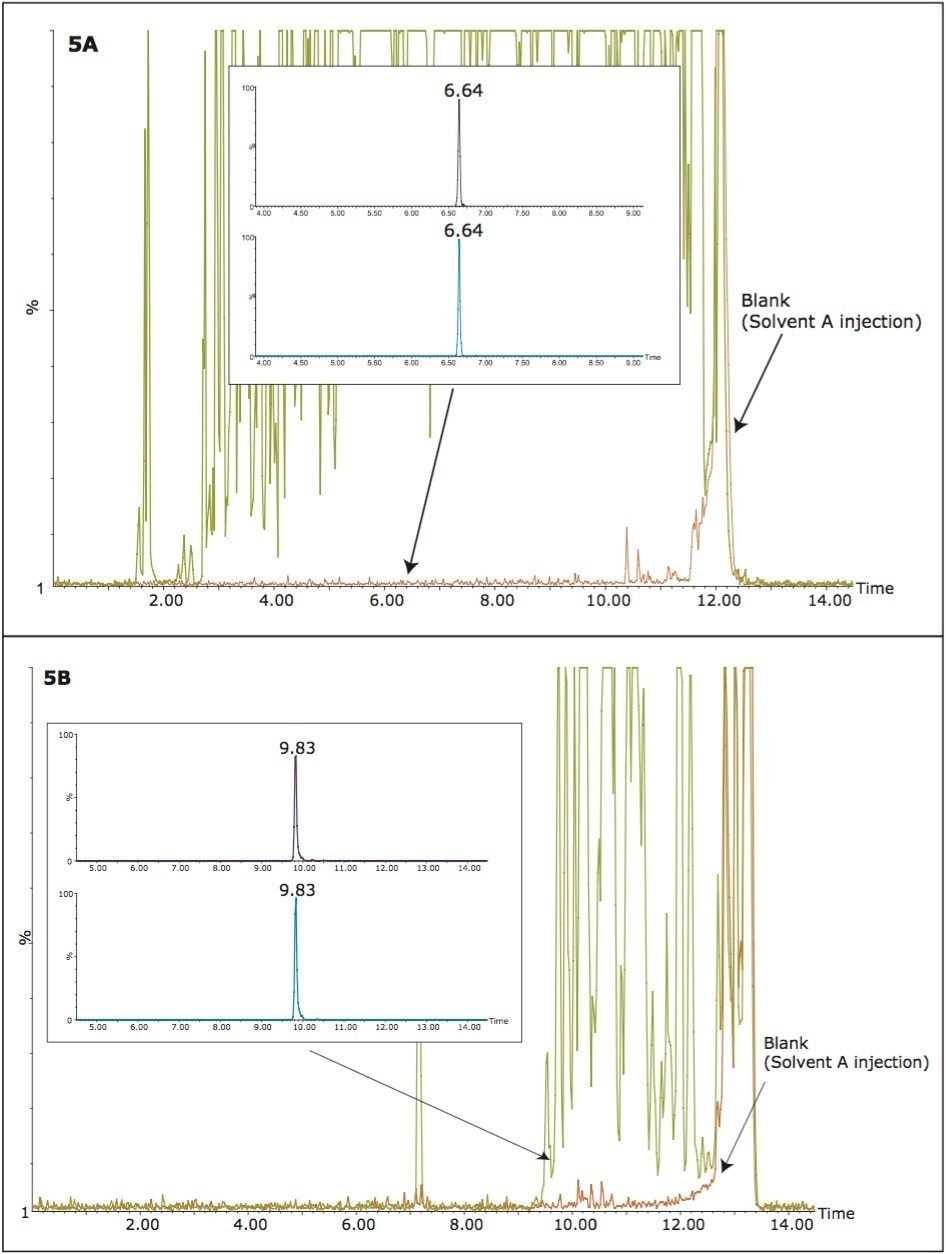
The increase in sensitivity of the 2D LC/MRM method is usually explained by the ability of extensive chromatographic separations that remove some of the co-eluting compounds responsible for analyte/IS suppression. To verify this hypothesis, the presence of background peptides from the serum digest was monitored by full scan MS scans (RADAR scans) during 1D- and 2D-MRM separations. These RADAR scans were scheduled throughout the chromatographic run (1 s scans), and they were performed at the end of each MRM cycle containing the four transitions (50 ms each) used for trastuzumab quantification. The data collected during these experiments are illustrated in Figure 5. During the 1D LC/MRM run, the chromatogram recorded with full scan MS acquisitions (RADAR chromatogram), shown in Figure 5A, reveals the true complexity of the serum digest sample. The inset shows the MRM chromatograms corresponding to the native/IS peptides. The signals of these peptides are significantly reduced (~four-fold) by the presence of the “heavy” peptide background contained in the SPE-cleaned sample. Figure 5B shows the RADAR chromatogram produced by the 2D high pH/low pH RP/RP chromatography with heart-cutting. The MRM chromatograms, shown in the inset, indicate that most of the background components that were co-eluting with the analyte in 1D chromatography no longer co-elute after two-dimensional chromatography. Data in Figure 5 provide a clear explanation for the increase in assay sensitivity observed in the 2D-MRM method.
720004510, December 2012