For research use only. Not for use in diagnostic procedures.
This is an Application Brief and does not contain a detailed Experimental section.
Low abundance proteins are often of biological interest and as such, sensitivity and low limits of quantification are key parameters in modern proteomic experiments. The SYNAPT G2-S System provides improved sensitivity, resulting in the routine detection of attomole levels of tryptically-digested proteins. In this application brief, we show results from a protein mixture spiked at different concentrations into a cytosolic E.coli lysate.
Comprehensively identify proteins at attomole concentrations in a complex biological matrix using SYNAPT G2-S.
Protein identification is often challenged by the sensitivity and specificity required. For example, the presence of contaminating peptides within the collision cell during the collision-induced dissociation process leading to mixed fragment ion spectra is often ignored. This is overcome by the use of mobility assisted data independent acquisition (HDMSE) to qualitatively and quantitatively characterize enzymatic protein digests. This method has proven to be highly efficient when dealing with protein mixtures of varying complexity. Advancements in instrument technology allow HDMSE data acquisitions to be accompanied with increased sensitivity by incorporating StepWave ion optics.
The StepWave device allows significantly more ions to be introduced while utilizing a robust mechanism for the elimination of neutral components and gas stream from the instrument, resulting in sub-femtomole sensitivity over a wide dynamic range. Here, we demonstrate the identification of known standard proteins spiked at varying levels into a complex sample matrix.
A standard protein mixture containing equal molar amounts of yeast enolase, bovine serum albumin, rabbit glycogen phosphorylase B, and yeast alcohol dehydrogenase, were spiked at various concentrations into 100 ng of digested E.coli cell lysate. This provided a dilution series ranging from 100 amol to 10 fmol. A Waters nanoACQUITY UPLC System coupled with a SYNAPT G2-S were used to separate and analyze the tryptic peptides. The SYNAPT G2-S was operated in LC-HDMSE mode, whereby the collision energy was switched between low and elevated energy state during alternate scans and associate precursor and product ions by means of retention and drift time alignment. Using ProteinLynx Global SERVER, data were processed and searched against a non-redundant E.coli database, with the four known protein sequences added and further correlation made that considered the physiochemical properties of peptides when they undergo fragmentation.
An example of an elevated energy spectrum at the 100 amol level is shown in Figure 1, which represents the ADH peptide ANELLINVK.
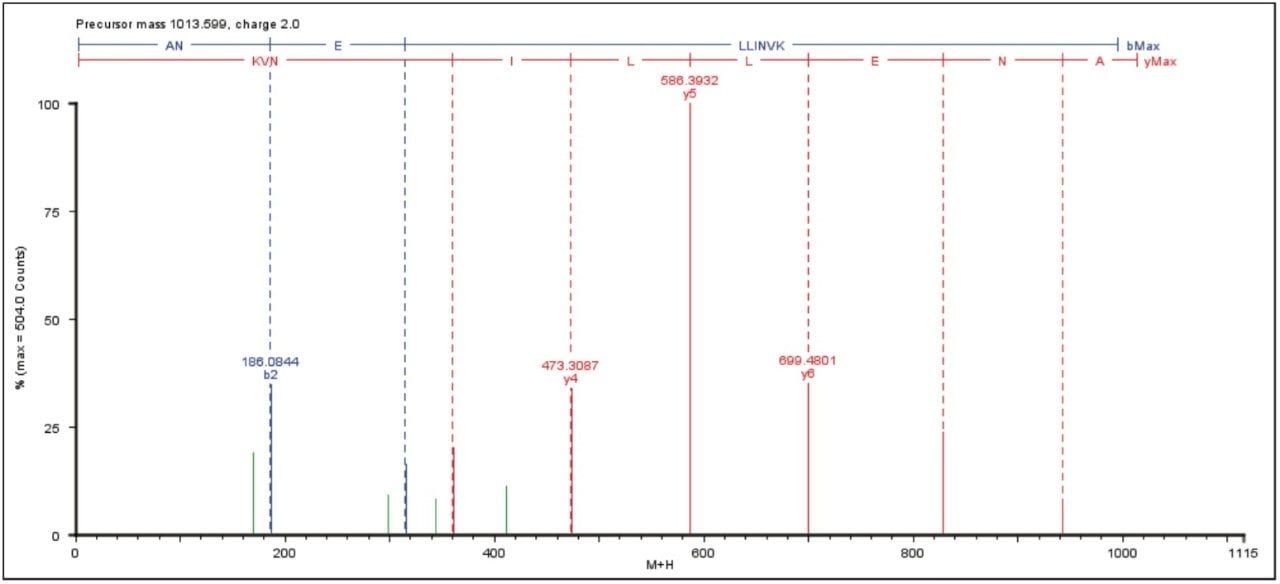
Figure 2 shows the average result of two technical replicates, illustrating the number of identified peptides, amino acid sequence coverage, and the intensity of the most abundant peptide identified to the proteins of interest.
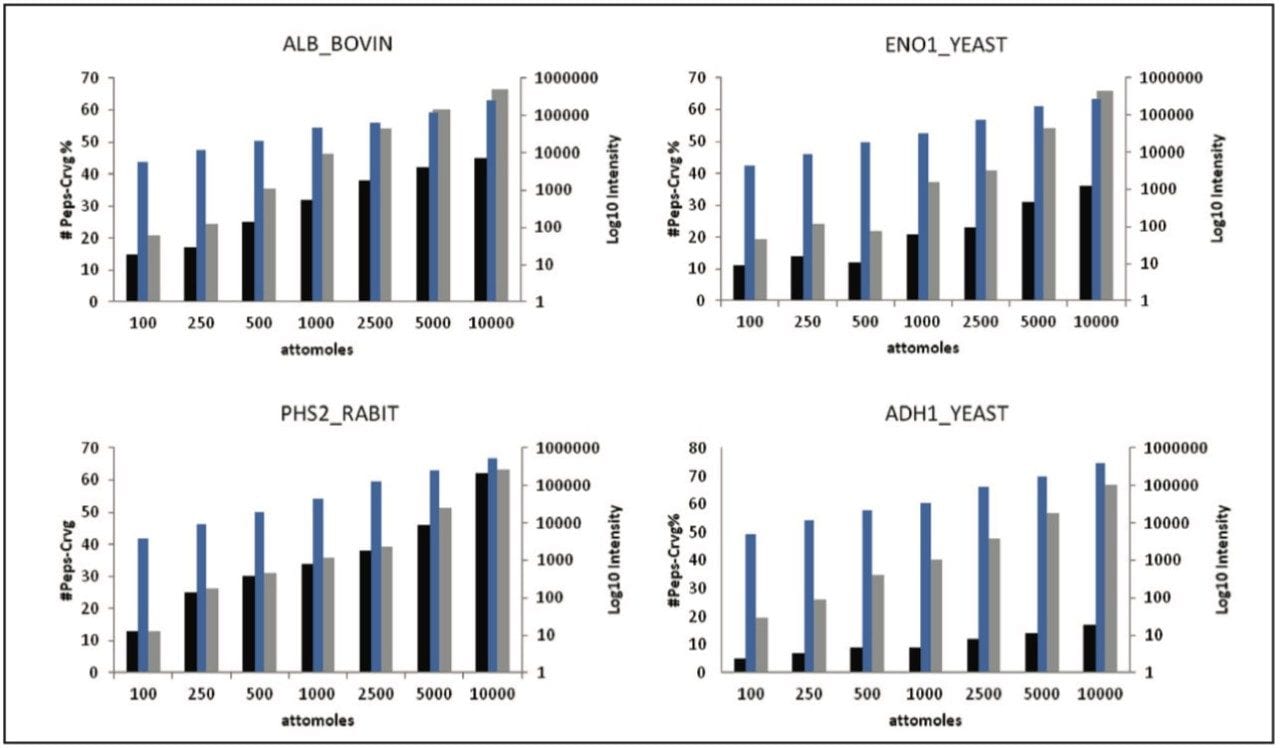
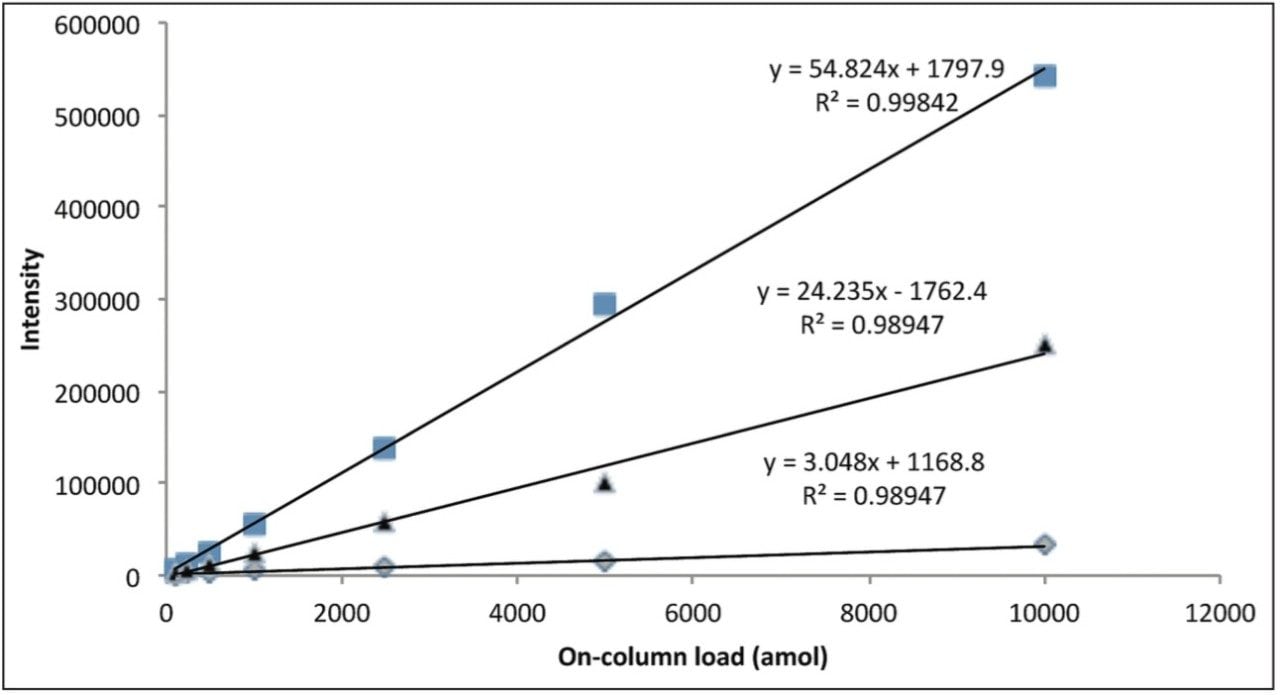
Figure 3 represents the linear response provided by three BSA peptides over the concentration range of the dilution series.
Here we have shown identification of four standard proteins spiked at varying concentrations (100 amol to 10 fmol) into a complex matrix using the SYNAPT G2-S System.
720004194, January 2012