In this application note, we describe a rapid and highly sensitive method for the routine analysis of RNAi duplexes preventing protein translation using the Waters ACQUITY UPLC System with Oligonucleotide Separation Technology (OST) Column chemistry.
UPLC method offers superior resolution, allowing for the detection and quantitation of excess single-stranded RNA, mismatched duplexes, and synthetic impurities.
RNA interference (RNAi) is a rapidly emerging strategy for temporarily silencing genes and preventing protein translation. RNAi is a double-stranded non-coding RNA molecule designed to bind to a specific mRNA target, and via a cascade of biochemical reactions interfere with the protein production. This method of gene silencing is currently being utilized in a variety of animal studies and is receiving increased of attention as a potential therapeutic strategy for humans.
A major challenge for developing human therapeutics preventing protein translation remains the assurance of RNAi purity. The presence of certain related impurities may lead to the possibility of unwanted, and perhaps detrimental, non-targeted gene silencing.
Major sources of impurities in RNAi duplexes are often the result of degradation, intra-molecular hybridization mismatches, or more commonly incomplete syntheses of the complementary single-stranded RNA complementary strands. The presence of non-hybridized single stranded RNAi is also undesirable and often associated with a decrease in therapeutic potency.
In this application note, we describe a rapid and highly sensitive method for the routine analysis of RNAi duplexes preventing protein translation using the Waters ACQUITY UPLC System with Oligonucleotide Separation Technology (OST) Column chemistry.
|
LC system: |
Waters ACQUITY UPLC System |
|
Column: |
ACQUITY UPLC OST C18 2.1 x 50 mm, 1.7 μm |
|
Column temp.: |
20 °C |
|
Flow rate: |
0.2 mL/min |
|
Mobile phase A: |
0.1 M TEAA, pH 7.5 |
|
Mobile phase B: |
20% Acetonitrile in A |
|
Gradient: |
35 to 85% B in 10.0 min (1% ACN/min) |
|
Detection: |
ACQUITY UPLC PDA, 260 nm |
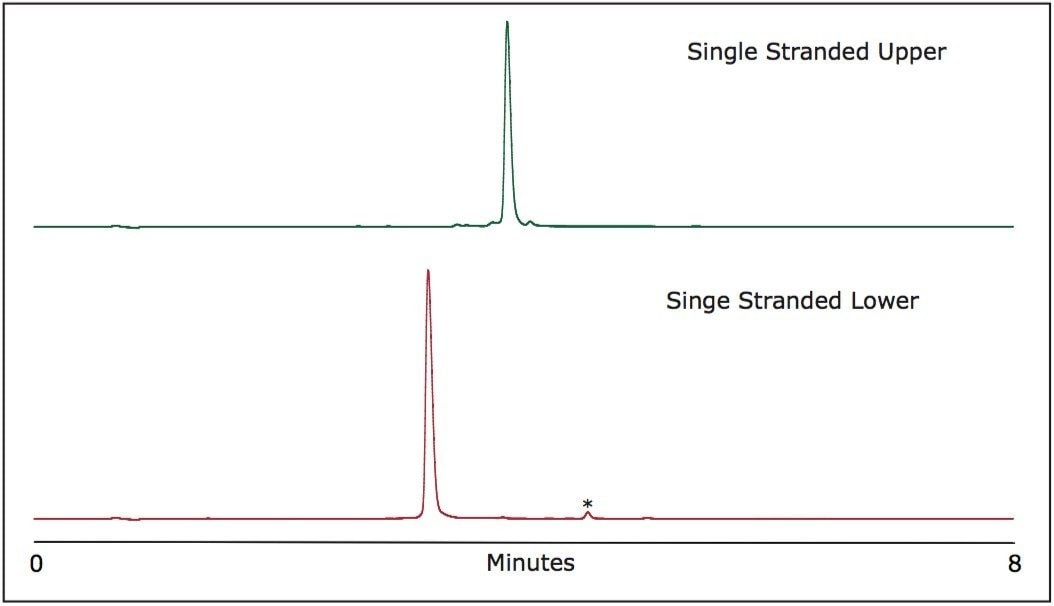
RNAi complementary strands 5’ - UCG UCA AGC GAU UAC AAG GTT - 3’ (upper) and 5’ - CCU UGU AAU CGC UUG ACG ATT - 3’ (lower) were purchased from Integrated DNA Technologies and reconstituted in 110 μL of 0.1 M triethylammonium acetate (TEAA) to yield concentrations of approximately 2.5 nmol/μL. The samples were purified prior to use1 and purity was verified prior to duplex formation experiments (Figure 1).
RNAi duplexes were prepared by combining appropriate molar ratios of upper and lower (2:1 and 1:2) complementary strands in 0.1 M TEAA. Mixtures were heated at 90 °C for 5 minutes and gradually cooled to 20 °C. Samples were prepared immediately prior to use to minimize sample degradation.
RNAi duplex samples were separated on a Waters ACQUITY UPLC System using an ACQUITY UPLC OST C18 2.1 x 50 mm, 1.7 μm column using ion-pairing reversed phase chromatography.2 Separated RNA species were detected with an ACQUITY UPLC PDA detector scanning from 19 to 350 nm.
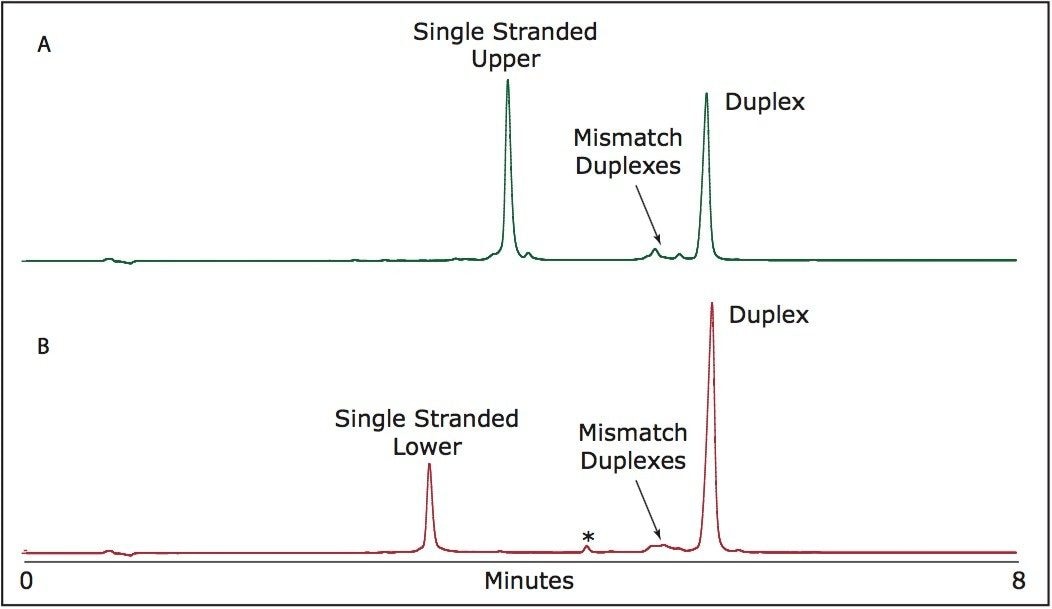
As shown in Figure 2, this system offered exemplary component resolution with no evidence of on-column duplex degradation or melting. Additionally, this method offers impressive separation of the desired duplex from impurities present in the sample, primarily mismatched sequences.
In the presence of excess single-stranded RNA, retention times of both the excess single-stranded RNA and duplex remain constant, highlighting the utility of this method for purification of RNAi. The method also allows for the separation of failure sequence and other mismatch duplexes from the desired duplex product.
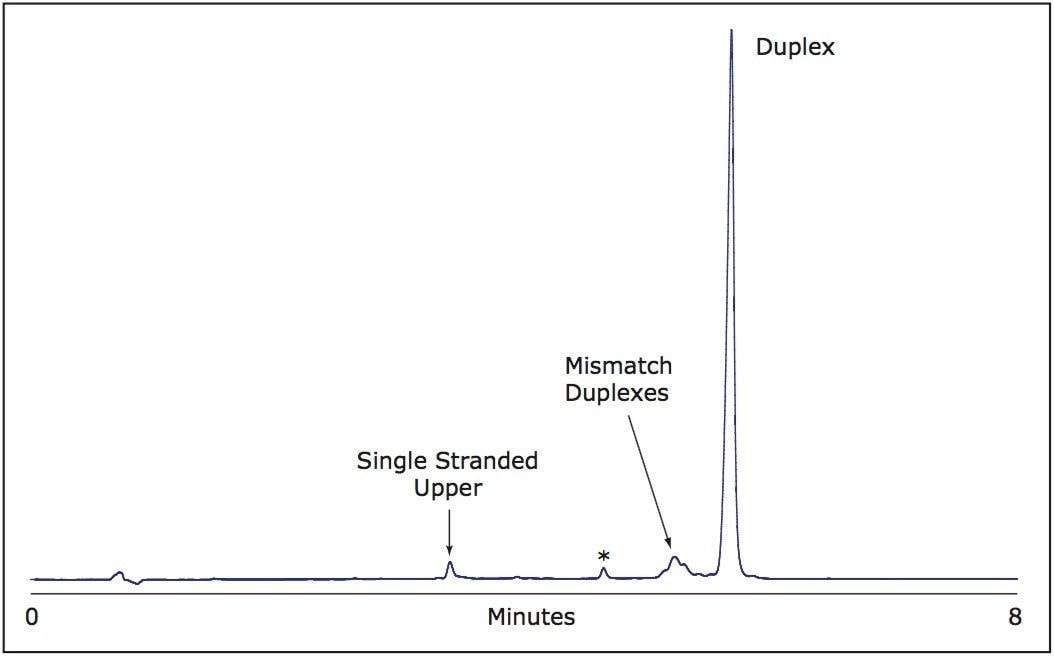
To fully investigate the utility of the chromatographic system and to confirm that there was no on-column melting of RNA duplexes with this method, we investigated whether our separation conditions would allow for the formation of RNA duplexes on an ACQUITY UPLC OST Column. To accomplish this, we separately injected each of the complementary RNAi strands under 100% binding conditions followed by gradient elution of the bound samples.
We found quantitative formation of an RNAi duplex following injection of upper and lower RNAi strands, with the resulting duplex peak exhibiting the identical retention time of authentic RNAi prepared via a separate annealing step (Figure 3). This data strongly indicates that under our UPLC separation conditions RNAi duplex does not melt during LC separation. In fact, it appears that spontaneous on-column annealing is favored as is indicated by the appearance of only one single-stranded peak present in a duplex/single-strand mixture.
Once quantitative duplex formation is accomplished, the duplex peak is predominant and well-separated from other impurities.
The data presented highlights the superior chromatographic resolution possible using the ACQUITY UPLC System and OST Column chemistry. This UPLC system solution offers superior performance for the efficient detection, quantification, and chromatographic resolution of RNAi duplexes from their single-stranded and mismatched counterparts, and also shows considerable utility in monitoring their formation.
The described UPLC method enables rapid and real-time quality control analysis of the reaction progress, eliminating the excessive RNAi characterization times associated with other analytical methods.
This method does not effect additional degradation of the duplex, enabling accurate, consistent, and reproducible analysis of RNAi duplexes. Finally, the UPLC method offers superior resolution, allowing for the detection and quantitation of excess single-stranded RNA, mismatched duplexes, and synthetic impurities.
Overall, this enables the direct analysis of reaction products and allows for purity determination in a high throughput manner.
720002573, April 2008