Long nucleic acids/transgene sequences in cell and gene therapies range in length from 500 to 10,000 nucleotides or more. Confirming their identity and purity is critically important to ensure the safety and efficacy of these new modalities. Waters chromatography columns and systems combined with optical detection and/or mass spectrometry provide powerful capabilities for confirming the physicochemical properties of these nucleic acids and of their fully assembled drug product; when packaged into lipid nanoparticles (LNPs), viral vectors (e.g. AAV), or virus-like-particles (VLPs) for cell delivery.
On-Demand Webinar: LC-MS CQA Analysis of RNA Therapeutics

Compliance-ready, networkable applications-based software to streamline LC-MS data acquisition, processing, and reporting.
Accelerate the journey from sample analysis to decision-making with compliance-ready MAP Sequence, CONFIRM Sequence and INTACT Mass Apps for comprehensive nucleic acid product understanding.
Columns and analytical standards that deliver leading-edge LC and LC-MS performance for nucleic acids
Obtain exceptional ion pair reversed phase (IP-RP) resolution, sensitivity, and stability with Waters MaxPeak Premier Oligonucleotide BEH C18 Columns that are QC-tested with Waters MassPrep Oligonucleotide Standard to ensure lot-to-lot reproducibility.
Achieve intact separation of long-chain nucleic acids, including mRNA and their fully packaged drug product, with Waters GTxResolve SEC 450 Å, 1000 Å, and 2000 Å columns that separate nucleic acids and assembled drug products based on size and shape, enabling payload quantification, product integrity, and purity analyses.
Obtain orthogonal selectivity and ion pairing free LC options with Waters Gene-based Therapeutics (GTx) Columns, including weak, and strong anion exchange columns and amide-bonded BEH particle HILIC columns for an array of new method options.
Eliminate the need to prepare complex mixtures with Waters prepackaged lot-certified Oligonucleotide and Nucleic Acid Standards that deliver ease-of-use and peace-of-mind when running or troubleshooting important separation-based assays.
Observe 3x increased sequence coverage and longer RNA digestion products with unique masses for LC-MS analysis. RapiZyme RNases offer complementary cleavage properties and overlapping digest products for improved confidence in identification.
Avoid running gels and realize tremendous time savings/greater efficiency when confirming plasmid length and determining linearization efficiency - a game changer for In Vitro Transcription (IVT) labs.
Optimize your laboratory's productivity and success with Waters Global Services to maintain peak system performance, minimize down time, address application challenges, and support stringent compliance requirements.
Maximize resources and minimize risk with payment options from Waters Capital, including upgrading aging equipment, getting customized support, and bundling services into one monthly payment.

LC-UV-MS analysis of the HPRT sgRNA digested with recombinant RapiZyme Cusativin. Three independent recombinant enzyme batches of RapiZyme Cusativin were used to digest HPRT sgRNA, and the three UV chromatograms (black, blue, and red traces) are overlayed on top of each other, indicating highly reproducible digestion behavior.
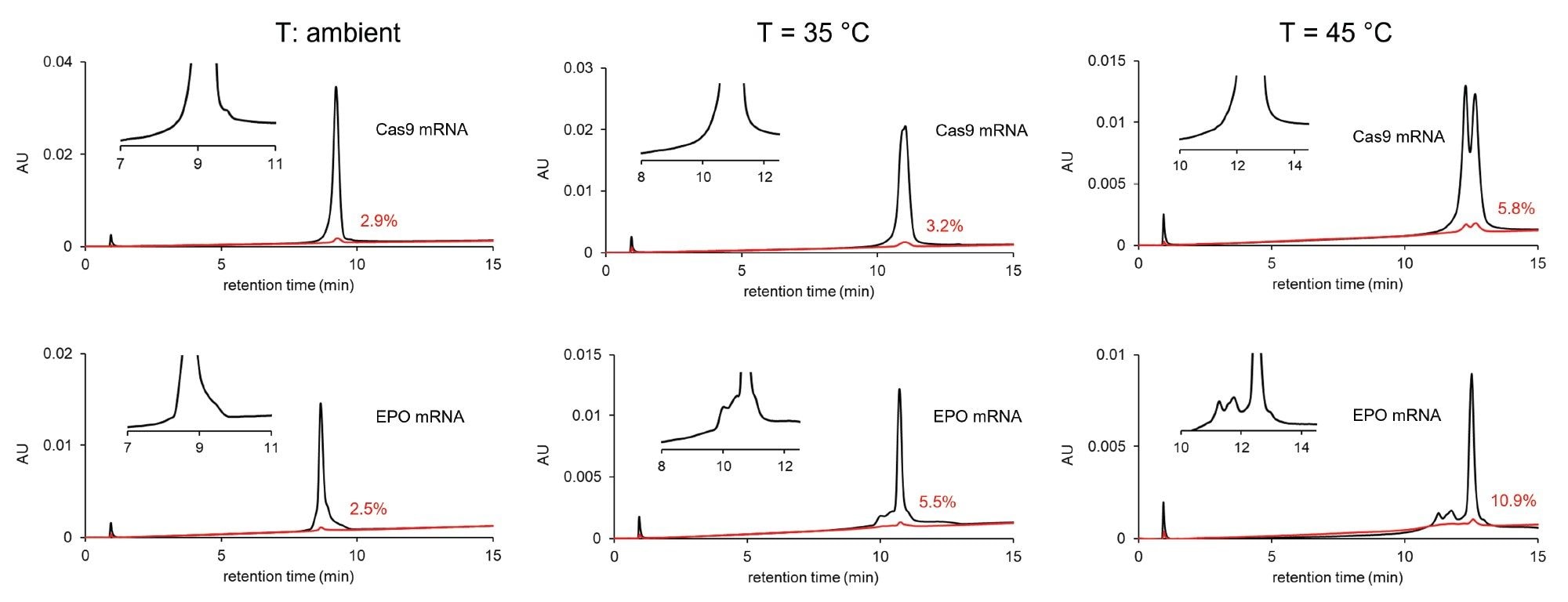
The effect of temperature on the chromatographic profile (selectivity) and carryover. Column: Gen-Pak FAX 100 x 4.6 mm, 2.5 µm Column, mobile phase A: 25 mM TRIS, pH=7.6, mobile phase B:25 mM TRIS, pH=7.6 + 2 M NaBr, F=0.6 mL/min, gradient: 12–35% B in 15 min (shallow gradient), modulator solvent plug: mobile phase B. Bracketing injection: 1 µL modulator pre-plug + 2 µL sample + 1 µL modulator post-plug. Temperature: ambient (left), 35 °C (middle) and 45 °C (right).
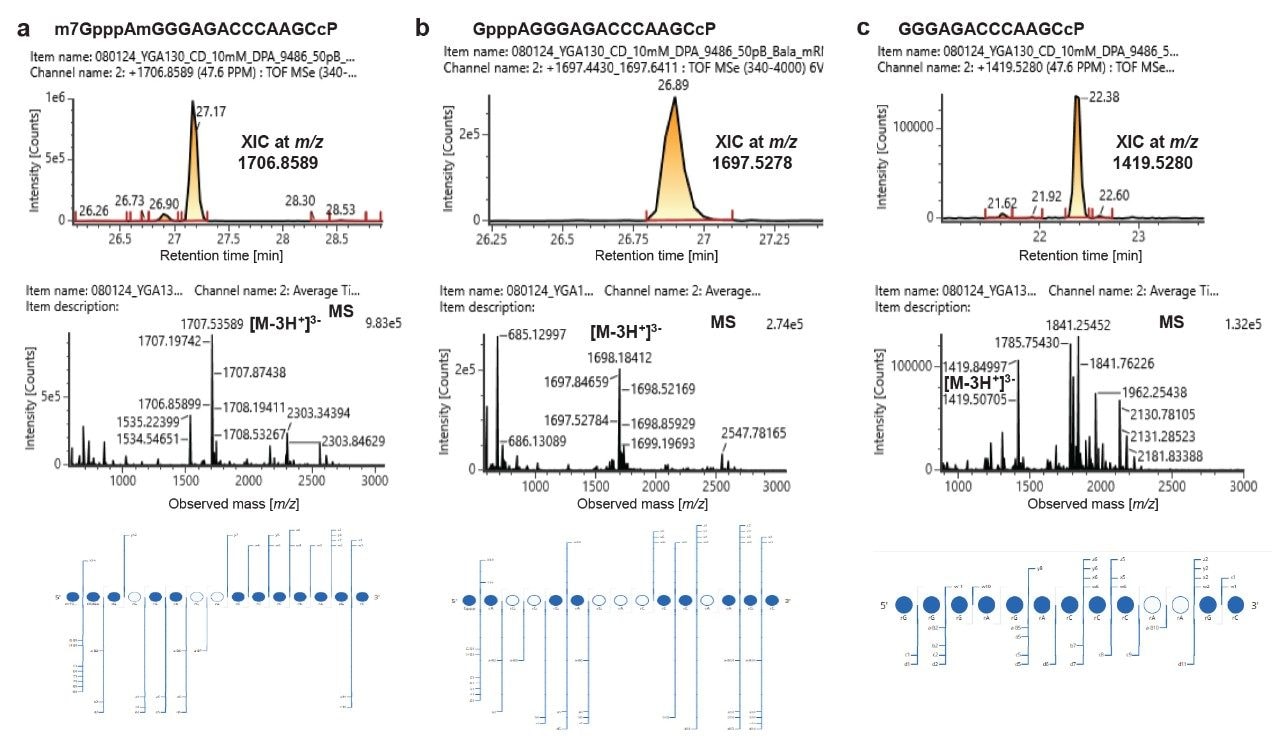
Characterization of different forms of 5’-capped oligonucleotides in the RNase MC1 digest of mRNA 1 including cap (a), unmethylated cap (b), and uncapped (c).
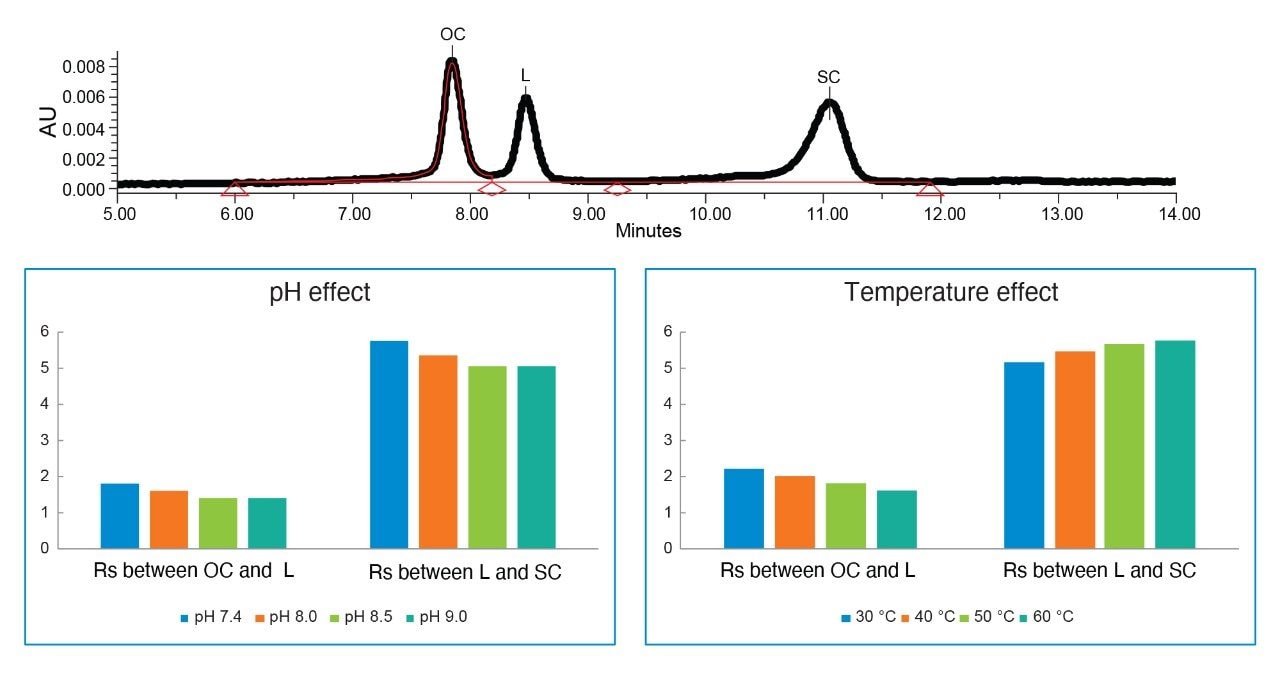
Effect of pH and temperature on the ΦX174 plasmid isoform separation.
