In this application note, several serum samples were analyzed using ion mobility enabled high resolution mass spectrometry. Data were processed using the software packages Mass-MetaSite and WebMetabase to automatically detect and visualize peptide catabolites. When combined with the accuracy and selectivity of HDMSE acquired data (ion mobility enhanced HRMS data independent acquisition), this allows the confident identification and tracking of catabolites.
Peptides have many attractive attributes as druggable compounds, but frequently suffer from poor pharmacological properties, such as permeability and stability, which make their development challenging. To overcome these limitations, peptides are being increasingly engineered or enhanced through the introduction of more complex structures and non-native modifications.
Identifying catabolites of these peptides is an important function of the drug development process, in order to understand clearance and metabolite fate, particularly in the context of unique human catabolites. Confident identification is analytically and computationally challenging; even simple sequences contain many hydrolyzable bonds that lead to hundreds of potential catabolites. For in vitro or in vivo studies, various amino acids or engineered peptide segments may also face additional transformative pathways such as oxidations, deamidations, conjugation and deconjuction. The multiply charged nature of peptides acquired using an electrospray based LC-MS analysis leads to a large number of m/z values, all of which need to be screened against to ensure that all catabolites are identified. All of these factors add complexity to peptide catabolite identification.
Somatostatin is a natural growth-inhibiting peptide hormone that has been the focus of numerous studies. Riera et al., published detailed descriptions of somatostatin and the influence of several sequence modifications on stability in human serum.3 In this application note, several of these serum samples were reanalyzed using ion mobility enabled high resolution mass spectrometry.
Data were processed using the software packages Mass-MetaSite and WebMetabase (Molecular Discovery Ltd.) to automatically detect and visualize peptide catabolites. When combined with the accuracy and selectivity of HDMSE acquired data (ion mobility enhanced HRMS data independent acquisition), this allows the confident identification and tracking of catabolites.4
Studies were analyzed using a centralized server running WebMetabase based in the UK, enabling multiple global users and labs in both the UK and USA to upload, process, view data simultaneously, and serve as a single repository for datasets. Additionally, functionality in Webmetabase enabling HELM notation (Pistoia Alliance) proved useful for visualizing complex peptide catabolite and fragment identification.
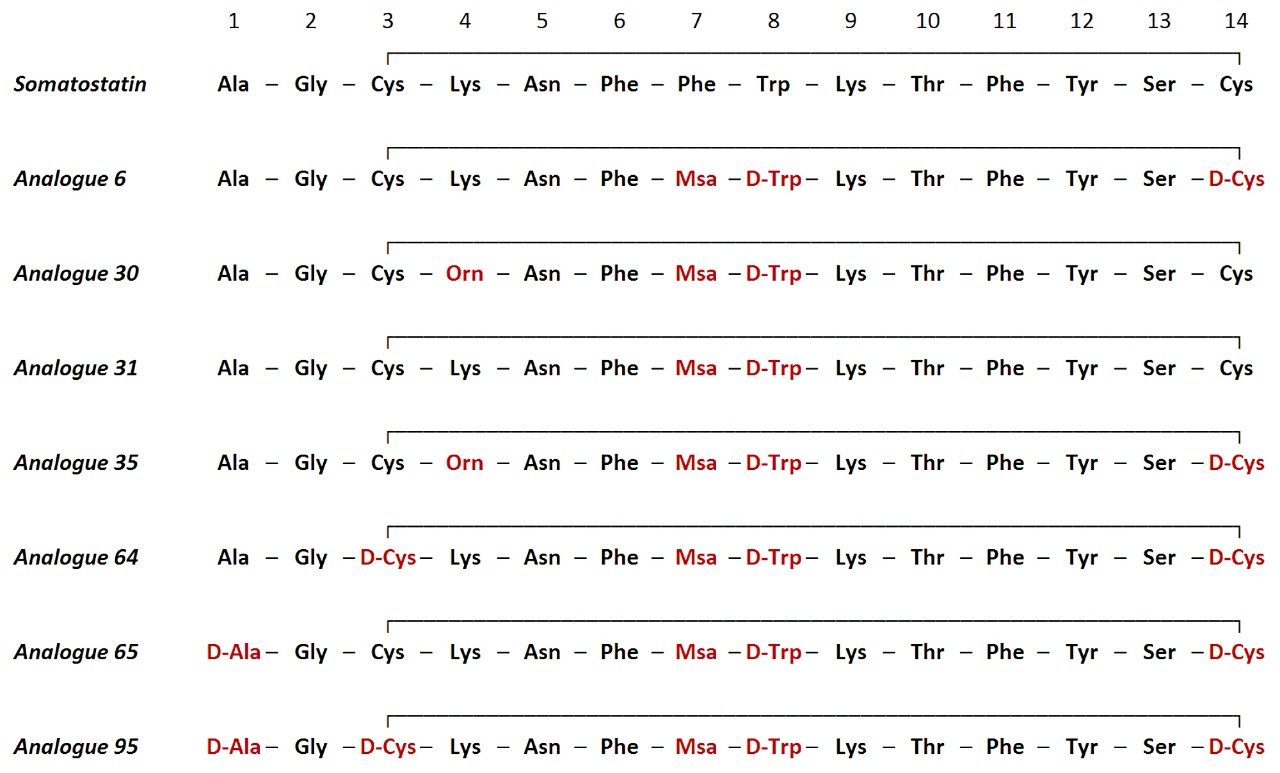
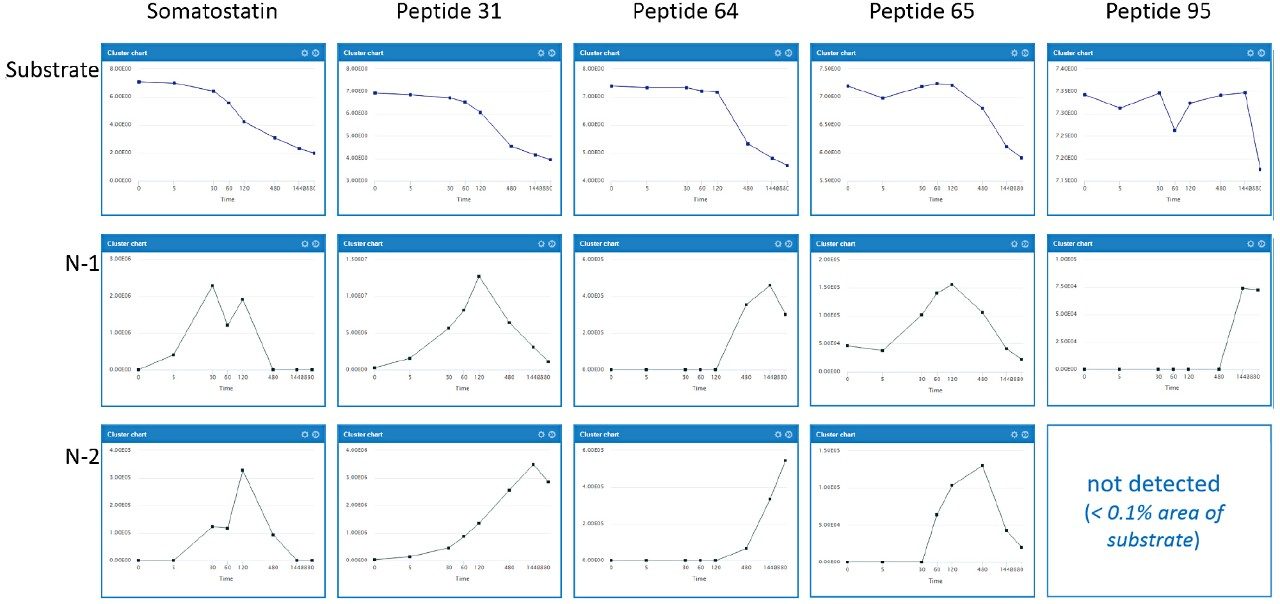
Eight 14-amino acid analogues of the hormone somatostatin were studied (Figure 1). Compounds were incubated in human serum at 11 time points (0 min, 5 min, 10 min, 30 min, 1 h, 2 h, 4 h, 8 h, 24 h, 30 h and 48 h) and evaluated for stability and major metabolite formation. Samples were kindly supplied by the laboratory of Professor Antoni Riera at the Institute of Research in Biomedicine, Institute of Science and Technology in Barcelona, Spain.
|
System: |
ACQUITY UPLC I-Class |
|
Vial: |
Polypropylene vials, 300 μL [p/n: 186002628] |
|
Column: |
ACQUITY CSH C18, 1.7 μm, 2.1 × 100 mm [p/n: 186005297] |
|
Column temp.: |
45 °C |
|
Sample temp.: |
8 °C |
|
Injection volume: |
1 μL |
|
Flow rate: |
0.4 mL/min |
|
Mobile phase A: |
water + 0.1% formic acid |
|
Mobile phase B: |
acetonitrile + 0.1% formic acid |
|
Gradient: |
2% to 30% B over 7 min, to 100% at 8 min, hold until 10 min |
|
System: |
Vion IMS QTof Mass Spectrometer |
|
Ionization mode: |
ESI positive ion mode |
|
Acquistion mode: |
HDMSE (with CCS) |
|
Acquisition range: |
m/z 50–2000 |
|
Capillary voltage: |
1.0 kV |
|
Collision energy: |
Low Energy 6 eV and High Energy 15–55 eV ramp |
|
Scan time: |
0.2 sec |
UNIFI 1.9.4 with API (application programming interface) enabled
Mass-MetaSite 3.4.2, WebMetabase 4.0 (Molecular Discovery Ltd.)
General data, TEC (two energies of collision) mode enabled
Metabolites generation, reactions monitored: Amide hydrolysis, oxidative deamidation, disulfide reduction
Mass, MS peaks, pattern filtering, tolerance (%): 30
Mass, Met ID, metabolite generations: 2
Mass, Met ID, substrate bond breaking limit: 1
Mass, Met ID, break metabolites: True, 1
TEC, mass spectrometer: Waters QTof Mass Spectrometer
TEC, algorithm thresholds: Chromatogram/MS/MSMS filtering: 0.95/0.95/0.95
TEC, signal threshold: 40
External LC-MS File converter: Version 2.1.9 UNIFI msvc64
HDMSE data were collected on a Vion IMS QTof Mass Spectrometer running UNIFI with fully integrated and automatic CCS reporting. Data were processed using Mass-MetaSite and uploaded using MetaSite Batch Processor to WebMetabase for visualization.1
All analogues studied contained substitution of Phe[7] by Msa and Trp[8] by D-Trp. Various permutations of Ala[1], Cys[3], and Cys[14] substitutions to their D-amino acid equivalents, as well as substitution of Lys[4] to ornithine, were assessed for stability. Across all analogues, WebMetabase was used to monitor disappearance of the substrates, evaluate the modulation of key metabolite formation (particularly -Ala and -AlaGly), and monitor for other major metabolic pathways, including ring opening. As previously reported, metabolism of native somatostatin primarily showed the loss of Ala (-71 Da) and -AlaGly (-128 Da) from the tail portion of the molecule.2,3
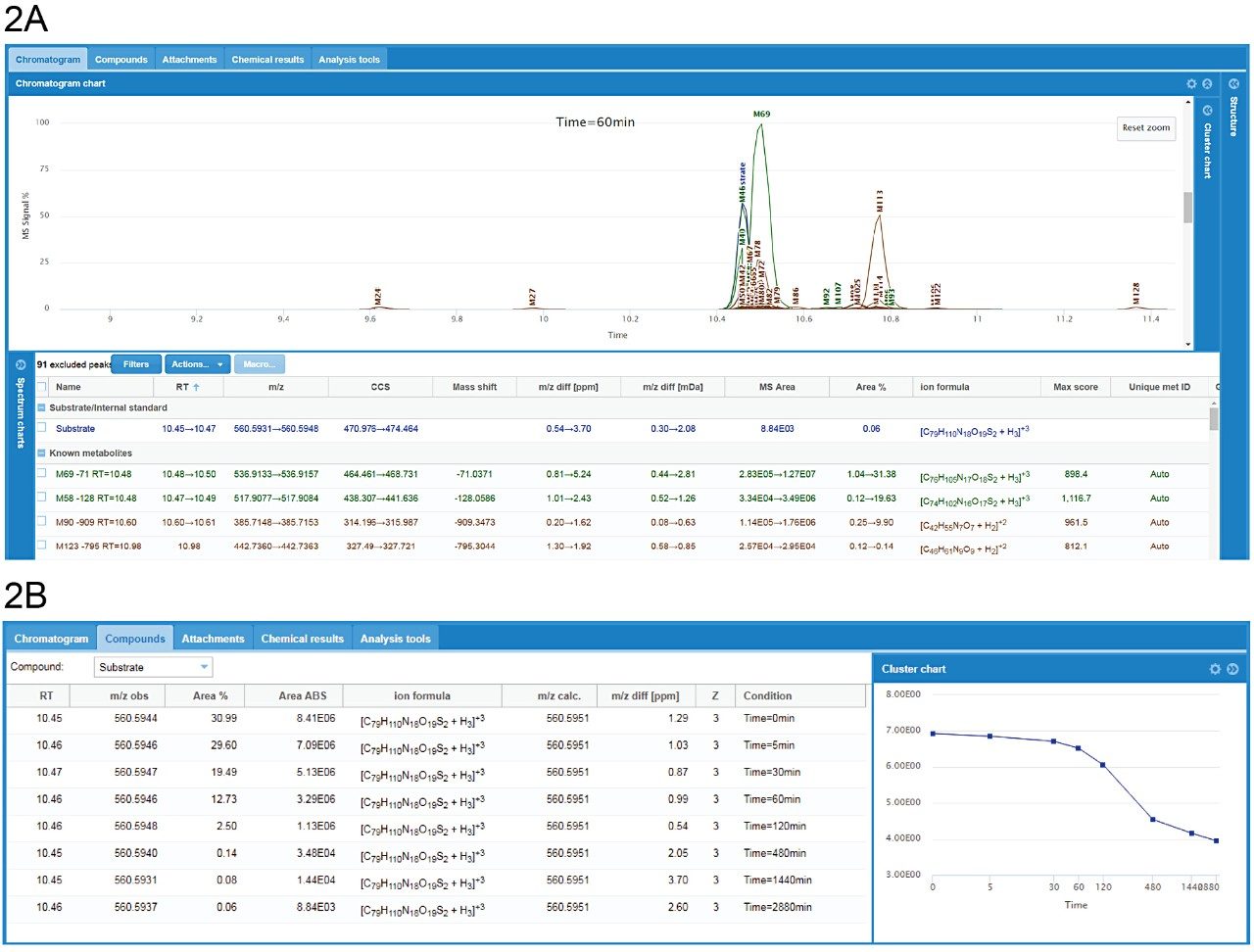
A representative example in WebMetabase, peptide 31, was largely cleared from the incubations over the 48-hour time course. By switching between the Chromatogram overview (Figure 2A) and Compounds overview (Figure 2B), the user can access summed chromatogram overlays, RT values, m/z values, mass differences from parent substrate, MS areas/responses, as well as % of substrate measured range for either the entire sample set or individual species across components. CCS values are also integrated directly in both Mass-MetaSite and WebMetabase. Ion mobility enables spectral cleanup, removal of non-related ions, and ensures that tracked peaks have not only the same m/z, but also CCS. This is especially beneficial inscenarios where peak shifting or closely luting isobaric peaks occur.
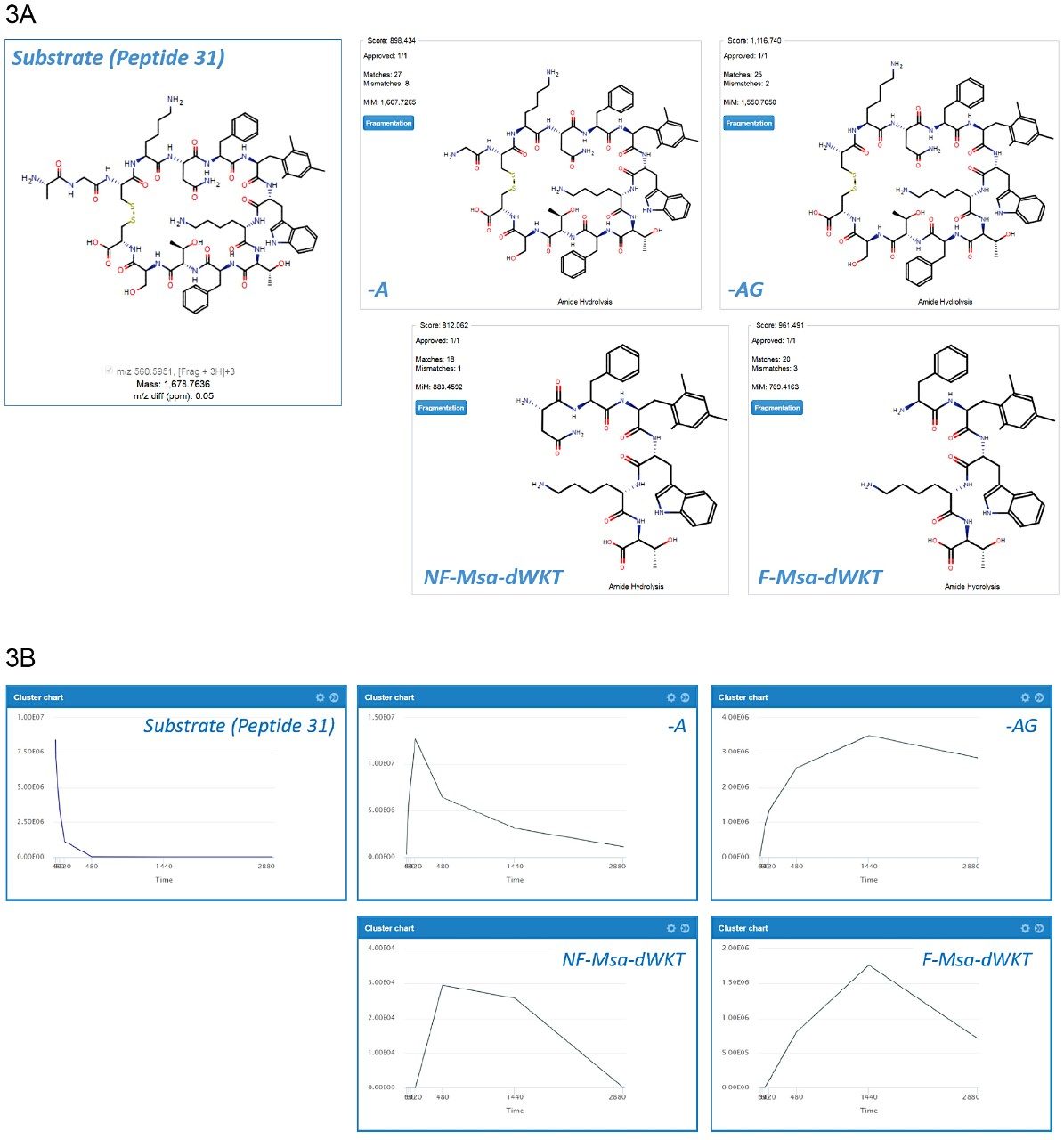
WebMetabase provides a visual representation for all metabolites detected, along with scoring algorithms to provide an assessment of all identified metabolites. Major metabolites and their structures are highlighted in Figure 3A. Scores for metabolite predictions are shown in the top left of each structure (all > 800) and the number of corresponding matched and mismatched spectral identifications are also provided. Figure 3B shows the response-time profiles for those compounds. The formation of -71 (-Ala) and -128 (-AlaGly) metabolites can be seen after 10 min and reaches a maximum level between 30 and 60 min. Subsequent ring opening products Asn-Phe-Msa-D-Trp-Lys-Thr and Phe-Msa-D-Trp-Lys-Thr (abbreviated NF-Msa-dWKT and F-MSa-dWKT, respectively) can be seen forming after 60 min.
Somatostatin response, along with four Msa/D-Trp modified peptides (31, 64, 65, and 95), were plotted for all substrate and N-1, N-2 catabolite losses in a detailed comparison (Figure 4). The majority of metabolism could be attributed to the N-1 and N-2 amino acid losses (-Ala and -AlaGly). Large differences in clearance were observed when the terminal alanine or bridging cysteine molecules were modified. The time/response profiles of the substrate, along with the primary N-1 and N-2 metabolites show the disappearance of the drug and how this relates to the appearance (or absence) of the major metabolites (Figure 4).
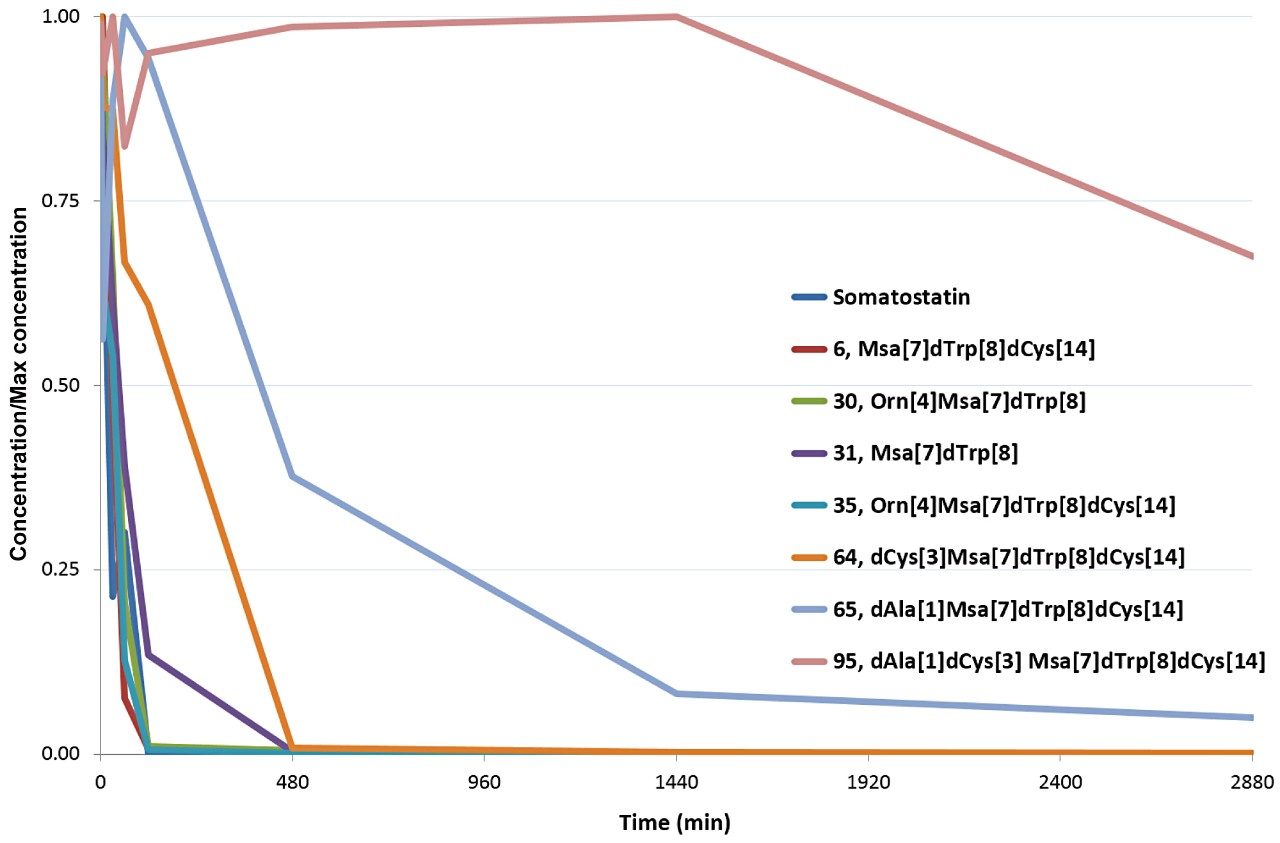
Once all peptide analogues were evaluated using WebMetabase, stabilities were plotted (in Excel) to show relative comparisons over two days (Figure 5). Catabolite information processed by WebMetabase showed that D-amino acid substitutions for Ala[1], Cys[3], and Cys[14] not only enhanced stability individually, but appeared to be synergistic. Some modifications (notably Cys[14] to D-Cys[14]) did not on their own have a noticeable impact, as in peptide 31 to peptide 6, but in combination with D-Cys[3] appeared extremely effective, as in peptide 6 to peptide 64, or peptide 65 to peptide 95. Lys[4] to Orn[4] and Phe[7]Trp[8] to Msa[7]D-Trp[8] substitutions showed modest differences relative to native somatostatin with respect to overall stability (detailed data not shown). Overall, peptide 95 appears very stable, which is further evidenced by the delayed and reduced appearance of any metabolism as compared to the other analogues.
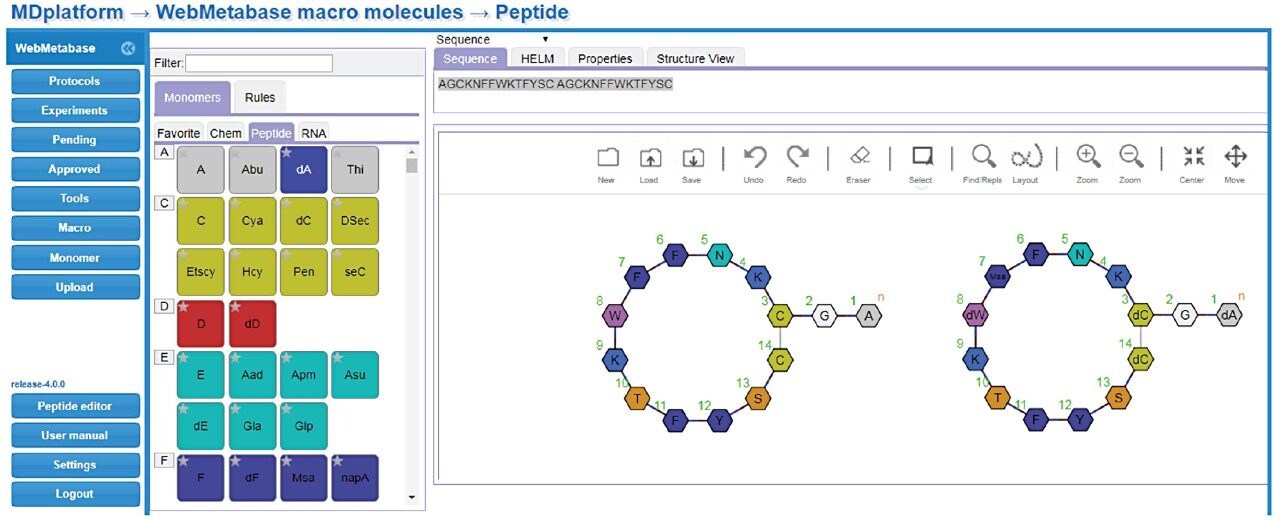
HELM notation enables viewing of complex macromolecular structures in a monomer format (Pistoia Alliance, https://www.pistoiaalliance.org/projects/current-projects/helm/). This feature retains chemical structure information for monomers and bridges the gap between sequence based structural notation and complex molecular drawings (e.g., .mol or .SDF formats) for large structures. An example of this is shown in Figure 6, where the molecule structures for somatostatin and peptide 95 are shown using HELM notation.
WebMetabase was used to process and visualize data from all eight somatostatin analogues. Catabolism primarily occurred at the terminal amino acids outside the disulfide linked ring portion of the structure. Major metabolic routes and their rates were monitored by observing parent stability and monitoring the appearance of key cleavages. Substitution of the L-cysteine and terminal L-alanine residues resulted in the largest improvements to stability. Some changes were observed to be synergistic, such as coupling D-Cys[3] to D-Cys[14], which led to a significant boost in observed stability. Vion data processed by WebMetabase enabled efficient identification of these modifications and their effects on reducing somatostatin degradation, making it clear when specific cleavages were enhanced or blocked via substitution.
The use of ion mobility and generated CCS values improved data quality by providing an additional tracking property for each peptide, enhancing mobility filtered XICs and providing the capability to dig further into the CCS effects on molecule size for specific substitutions or cleavages. Ion mobility data processed using the Mass-MetaSite and WebMetabase delivered an efficient method to review complex peptide catabolism data in a straightforward manner.
The integration of functionality from third party vendors such as Molecular Discovery enabled rapid access to features such as IMS enhanced workflows, HELM integration, and cloud-based visualization packages. The utility of HELM integration facilitated a viewpoint more familiar to biologists (rather than .mol or .SDF formats), and will continue to be useful as more structurally challenging molecules and hybrids are developed. The ability to store, process, and share data in the cloud enabled the use of a centralized server (based in the UK) to be accessed by multiple project users based at several locations globally. Data was routinely shared across sites in order to discuss observations and harness the knowledge of several researchers. Browser-based access to WebMetabase enabled the facile viewing from multiple PCs and locations and reduced the need for expensive hardware for client PCs and the requirement to install local copies of software.
720006586, August 2019