For research use only. Not for use in diagnostic procedures.
Here, we analyzed urine-based samples collected from rats dosed with tienilic acid (TA), a loop diuretic, to demonstrate the benefits and throughput capabilities when using the combined approach of HILIC RAMMP with ion mobility mass spectrometry.
Large scale metabolic phenotyping studies require the ability to perform accurate and reproducible analysis, but when using current profiling LC-MS methods, large sample cohorts can take weeks to complete, often across several batches of analysis. Acquiring the data in these multiple batches can lead to variation in the reproducibility of the assay with differences between batches developing due to variation in signal intensities from day to day.1,2 Reducing the overall batch run time can greatly improve this reproducibility alongside increasing laboratory throughput. Typical LC-MS profiling assays have sample cycle times greater than 10 minutes and in particular, hydrophilic interaction liquid chromatography (HILIC) requires extended re-equilibration phases prior to subsequent sample injections. For example, a continuous analysis based on conventional UPLC for a study cohort of 1000 samples would require several days of instrumentation time. Previous studies utilizing rapid microbore metabolic profiling (RAMMP) have shown comparable group discrimination and improved selectivity over conventional UPLC chromatography.3 The number of overall detected features with RAMMP can be compromised when compared with UPLC. However, combining the RAMMP methodology with data independent acquisition (DIA) strategies involving an ion mobility (IMS) workflow, such as HDMSE,4 results in both high peak capacity and ultimately larger numbers of detected features. Here, we analyzed urine-based samples collected from rats dosed with tienilic acid (TA), a loop diuretic, to demonstrate the benefits and throughput capabilities when using the combined approach of HILIC RAMMP with ion mobility mass spectrometry.
Rat urine was collected from 16 rats dosed with tienilic acid, tienilic acid isomer, and a control dose solution of a 0.25M trizma base solution at two, six, and 24 hours. A pooled sample was prepared by combining 10 µL of all samples (vehicle and dosed) which was subsequently stored at -20 °C until use. Prior to analysis, each sample was diluted at a ratio of 1:10 with LC-MS grade deionized water.
|
LC system: |
ACQUITY UPLC I-Class |
|
Column: |
BEH Amide 1.7 μm, 1 mm Å~ 50 mm |
|
Column temp.: |
50 °C |
|
Sample temp.: |
4 °C |
|
Injection volume: |
0.2 μL |
|
Flow rate: |
0.2 mL/min |
|
Mobile phase A: |
0.1% formic acid in water |
|
Mobile phase B: |
0.1% formic acid in acetonitrile |
|
Time |
%B |
|---|---|
|
0 |
99 |
|
0.03 |
99 |
|
2.33 |
50 |
|
3.33 |
99 |
|
3.33 |
99 |
|
MS system: |
SYNAPT G2 Si |
|
Ionization mode: |
ESI +/- |
|
Acquisition range: |
50–1200 m/z |
|
Capillary voltage: |
2.5 kV |
|
Acquisition mode: |
HDMSE 50 to 1200 m/z (low and elevated energy) |
|
Cone voltage: |
35 V |
|
IMS T-wave velocity: |
700 m/z |
|
IMS T-wave pulse height: |
40 V |
|
Collision energy: |
Low energy function at 6 eV and elevated energy function 20–50 eV |
|
Resolution: |
30,000 FWHM |
|
MS software: |
MassLynx |
|
Informatics: |
Progenesis QI, EZInfo, TargetLynx |
The LC-MS metabolite data were processed and searched with Progenesis QI. Peak picking and normalized label-free quantification was achieved with additional multi-variate statistical analysis conducted using EZInfo (Umetrics, Sweden). Compound searches were conducted using a variety of database sources, including HMDB.
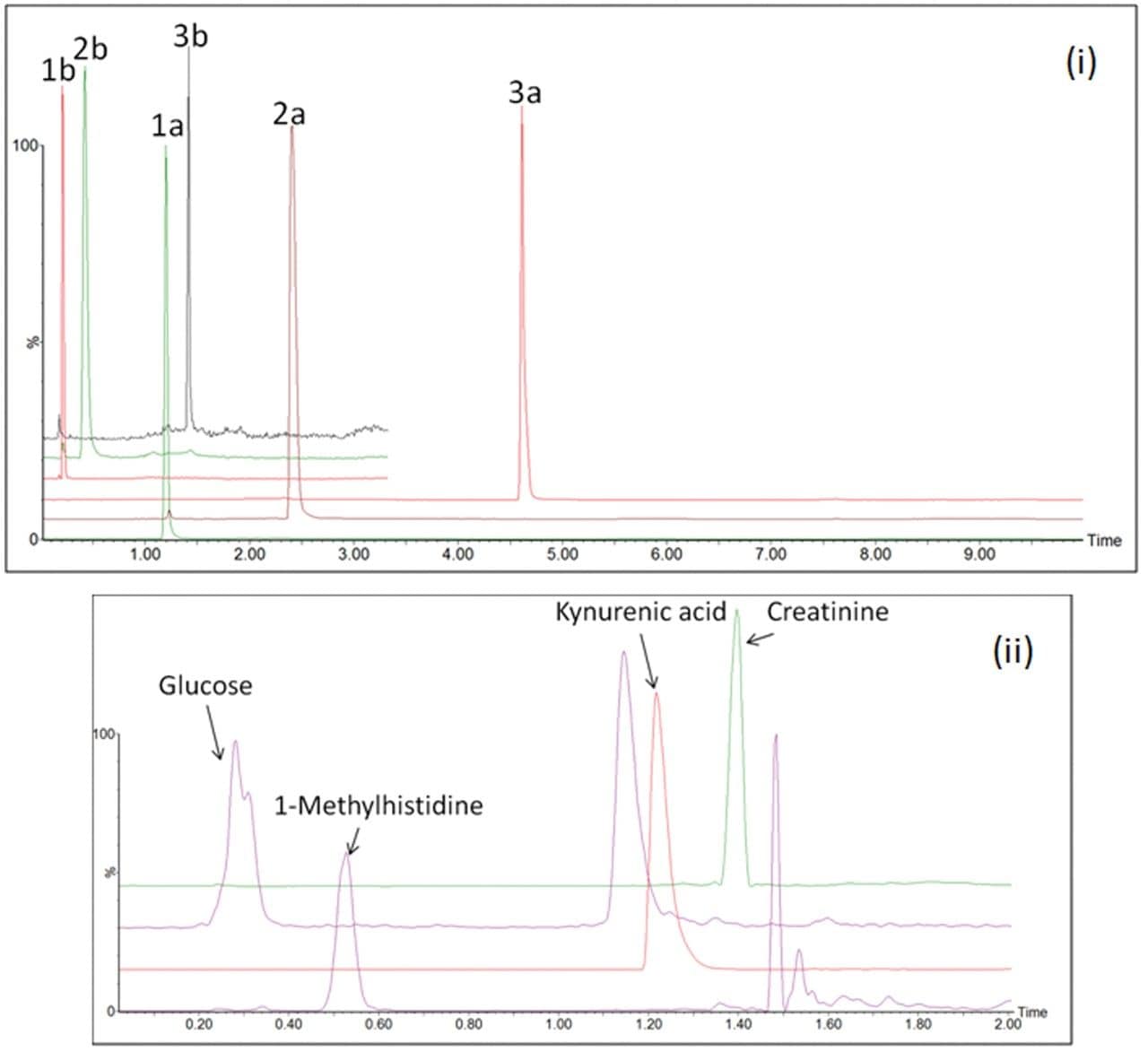
To address issues of batch effects across phenotyping studies, a reduction in the overall batch run time is required. Simply speeding up the chromatographic gradient and shortening the sample cycle time is not sufficient as this will ultimately affect the overall chromatographic performance, reducing the assays efficiency and fundamentally the reliability. The chromatographic method used in this study was scaled down from a conventional 10 min HILIC profiling method to a RAMMP method of 3.3 min which showed minimal impact on chromatographic separation. Figure 1(i) is an example chromatogram representing a LC-MS system suitability mixture, for comparison of conventional UPLC and RAMMP HILIC. Extracted ion chromatograms (XIC) for compounds of interest related to the study sample set include compounds such as glucose, 1-methylhistidine, kynurenic acid and creatinine, which are identified from the pooled QC (Figure 1(ii)) and demonstrate the utility of the RAMMP HILIC workflow.
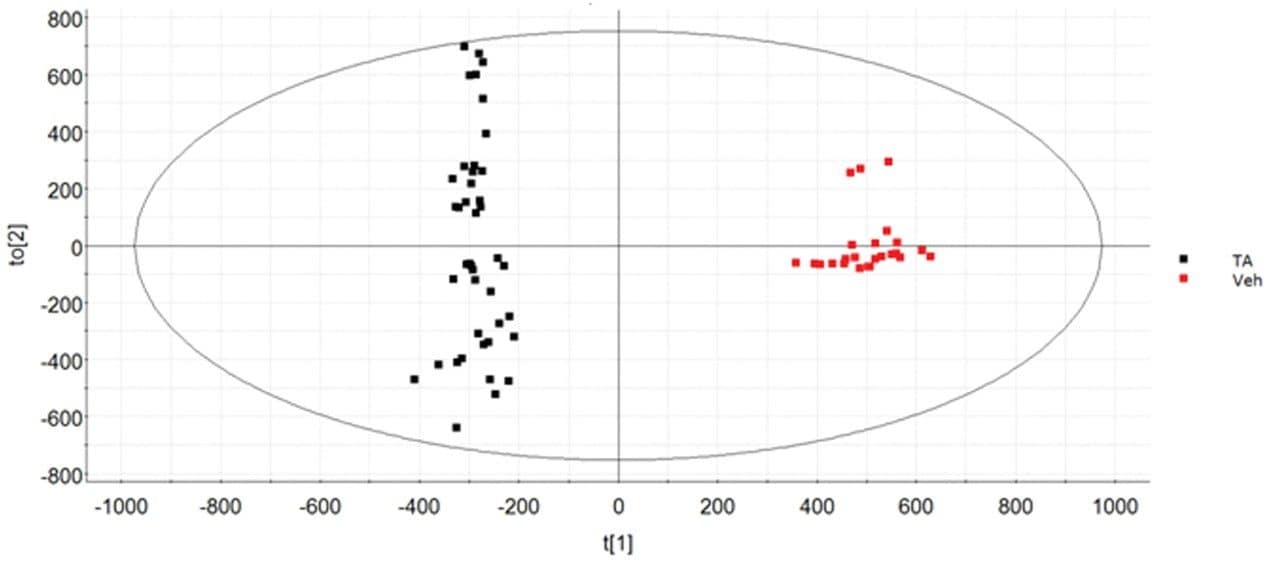
Multivariate statistical analysis (MVA) shows clear separation between vehicle and TA dosed rats, with discrimination between both groups demonstrated when applying an orthogonal partial least squares (OPLS-DA) approach (Figure 2). To ensure that the peak capacity is maintained when switching from conventional UPLC to RAMMP, IMS was implemented as part of the workflow resulting in an overall increased peak capacity of 51%.
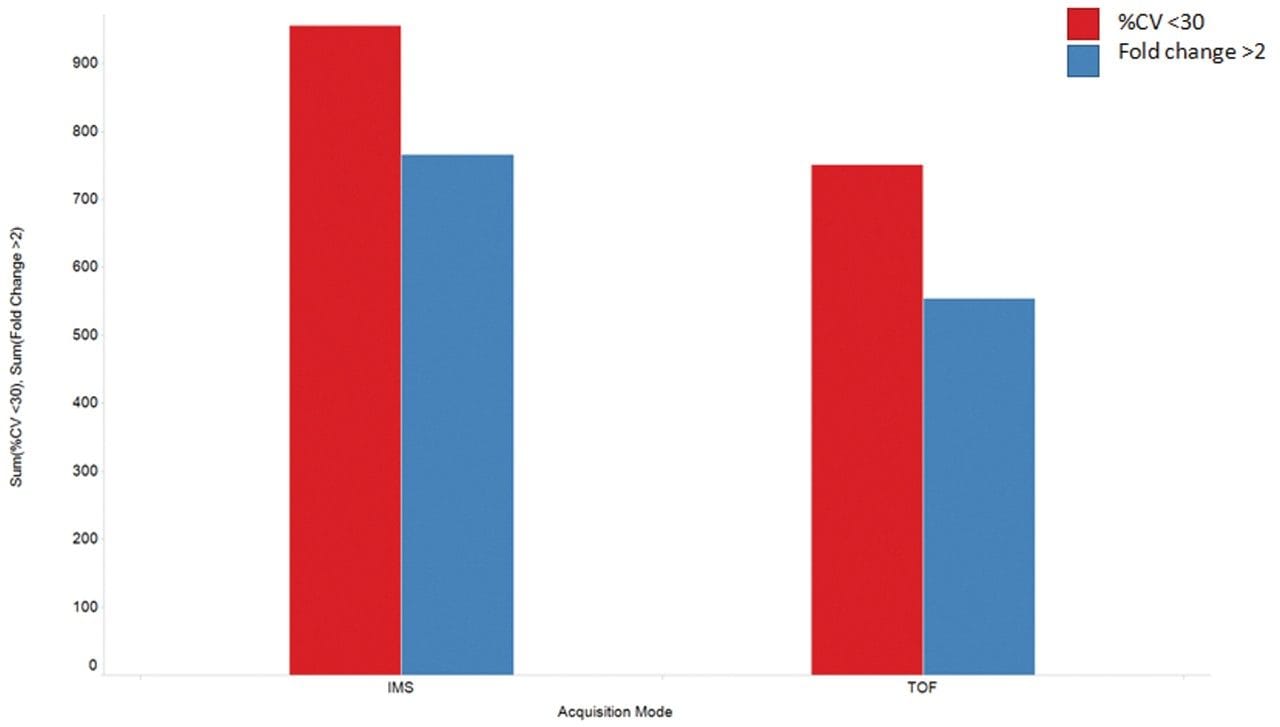
As demonstrated previously,5 this correlates with an increased number of detections when implementing IMS (Figure 3). A large proportion of the same statistically validated features are detected in both cases, however an additional 16% are identified as unique to IMS.
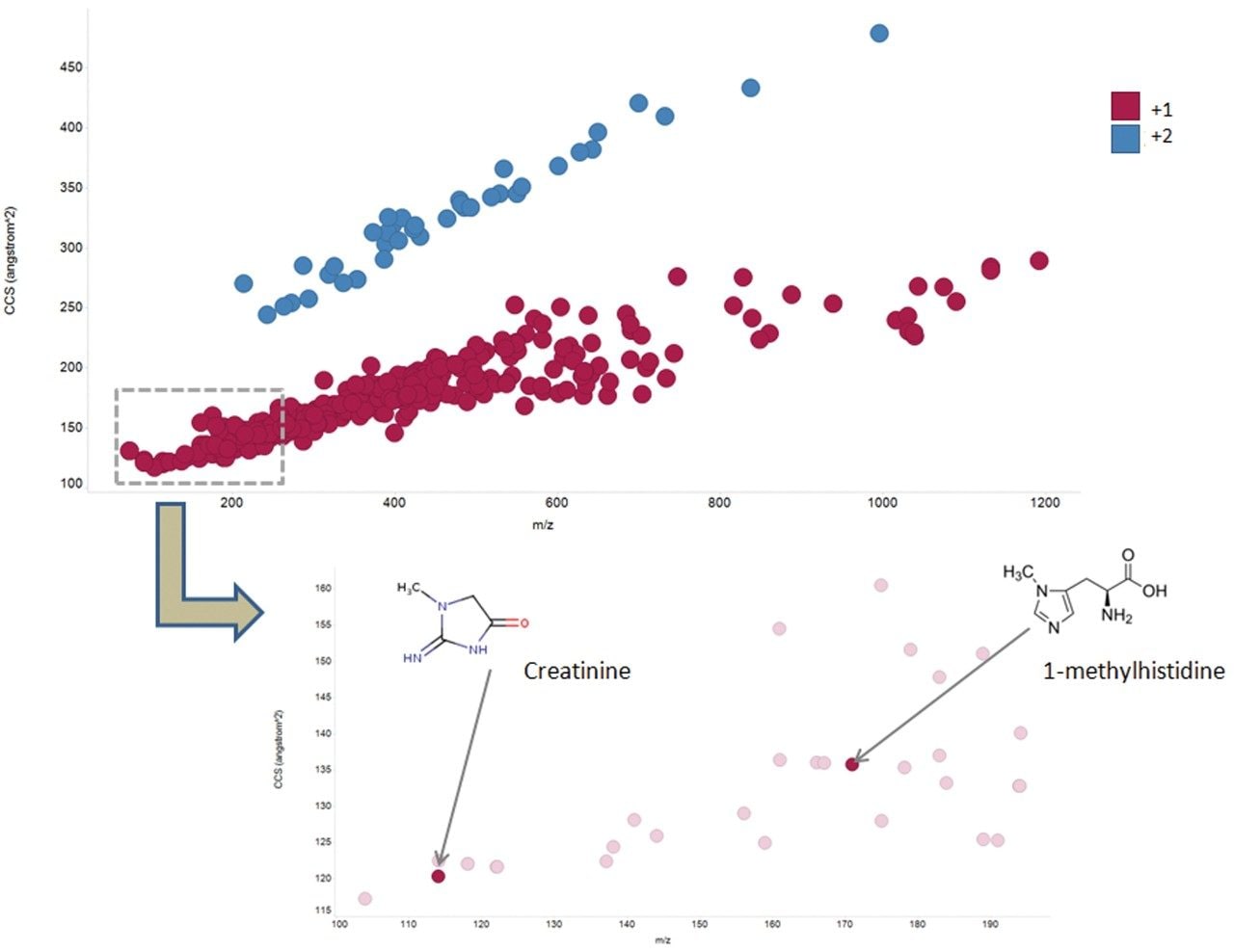
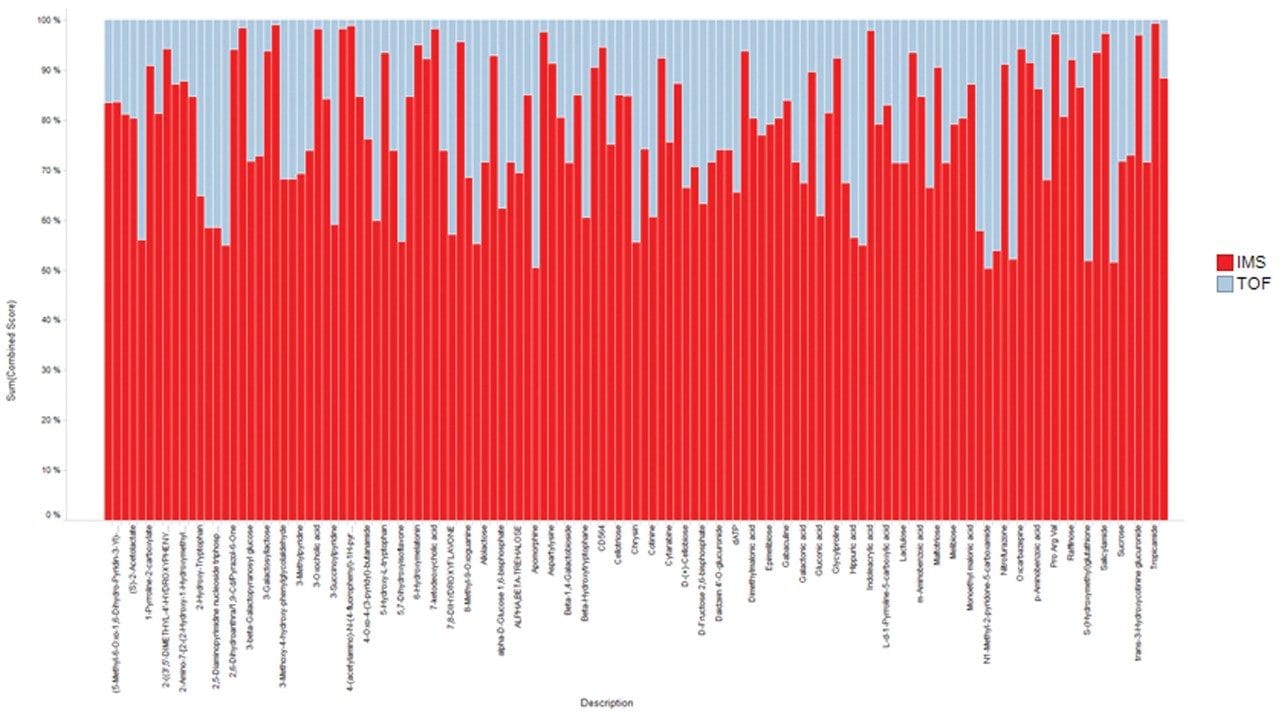
Additional benefits of implementing IMS also include improved spectral clarity and measurement of collision cross section (CCS) values. Figure 4 shows the CCS distribution with m/z for the QC pool highlighting both charge state separation and CCS determination of all detected features, including creatinine and 1-methylhistidine as example compounds. These experimentally determined CCS measurements are within 5% of the predicted CCS values.6 The ability to mobility separate and thereby increase peak capacity provides higher confidence results, containing fewer false positives. Based on the Progenesis QI processed results, cumulative identification scores are shown to have an average increase of 56% for the curated features of the IMS based dataset (Figure 5).
720006276, April 2018