A label-free, multi-omics approach provided qualitative and quantitative information in a single experiment to help characterize and investigate the effects of a glucosylceramide inhibitor for treatment of obesity. Progenesis QI and Progenesis QI for Proteomics Software enabled a seamless workflow, from LC-MS data processing and database searching to pathway interrogation, implicating carbohydrate metabolism, and molecular transport networks in the compound’s mechanism of action.
An increasing problem for human health, obesity is epidemic worldwide, affecting more than 500 million people. Obesity arises because of abnormal or excessive fat accumulation, a condition associated with a number of diseases, such as type 2 diabetes, heart and liver disease, and various cancers. Previous studies involving the treatment of mice with glucosylceramide inhibitors such as MZ-21 show reduced blood glucose levels and increased insulin sensitivity.1 To better understand the biochemical mechanism of action of such inhibitors in obese subjects, a multi-omic study involving protein and lipid analysis was conducted. Adopting a label-free, LC-HDMSE (LC-DIA-IM-MS) approach, the study provided qualitative and quantitative information from a single experiment. The curated data sets, interrogated using pathway-analysis tools, indicated that physiological processes such as hepatic-system development, inflammatory response, and carbohydrate metabolism are influenced following MZ-21 treatment.
|
LC system: |
nanoACQUITY UPLC |
|
Trap column: |
nanoACQUITY UPLC Symmetry C18, 5 μm, 180 μm x 20 mm (p/n 186007238) |
|
Column: |
ACQUITY UPLC M-Class HSS T3, 1.8 μm, 75 μm x 150 mm (p/n 186007473) |
|
Column temperature: |
35 °C |
|
Flow rate: |
300 nL/min |
|
Mobile phase A: |
0.1% formic acid in water |
|
Mobile phase B: |
Acetonitrile |
|
Gradient: |
3% to 40% B in 90 min |
|
Injection volume: |
1 μL |
|
LC system: |
ACQUITY UPLC |
|
Column: |
ACQUITY UPLC BEH C8, 1.7 μm, 2.1 mm x 100 mm (p/n 186002878) |
|
Column temperature: |
65 °C |
|
Flow rate: |
500 μL/min |
|
Mobile phase A: |
10 mM ammonium formate Acetonitrile-water (6:4) |
|
Mobile phase B: |
10 mM ammonium formate Isopropanol-acetonitrile (9:1) |
|
Gradient: |
30% to 99% B in 20 min |
|
Injection volume: |
3 μL |
|
MS system: |
SYNAPT G2-Si |
|
Ionization mode: |
ESI (+) at 3.2 kV |
|
Cone voltage: |
30 V |
|
Acquisition mode: |
HDMSE, 50 m/z to 2000 m/z, low-energy and elevated-energy functions |
|
Acquisition rate: |
0.5 s, low-energy and elevated-energy functions |
|
Collision energy: |
Low-energy function: 5 eV; elevated-energy-function: from 19 eV to 45 eV |
|
Resolution: |
25,000 FWHM |
|
IMS T-wave velocity: |
700 m/s |
|
IMS T-Wave pulse height: |
40 V |
|
MS system: |
Xevo G2-S |
|
Ionization mode: |
ESI (+) at 2.2 kV; ESI (-) at 1.7 kV |
|
Cone voltage: |
30 V |
|
Acquisition mode: |
MSE, 50 m/z to 2000 m/z, low-energy and elevated-energy functions |
|
Acquisition rate: |
0.1 s, low-energy and elevated-energy functions |
|
Collision energy: |
Low-energy function: 5 eV; elevated-energy function: from 20 eV to 45 eV |
|
Resolution: |
30,000 FWHM |
Progenesis QI
Progenesis QI for Proteomics
EZInfo (Umetrics, Umea, Sweden)
Spotfire (TIBCO Spotfire, Boston, MA)
Ingenuity Pathway Analysis (Qiagen, Redwood City, CA)
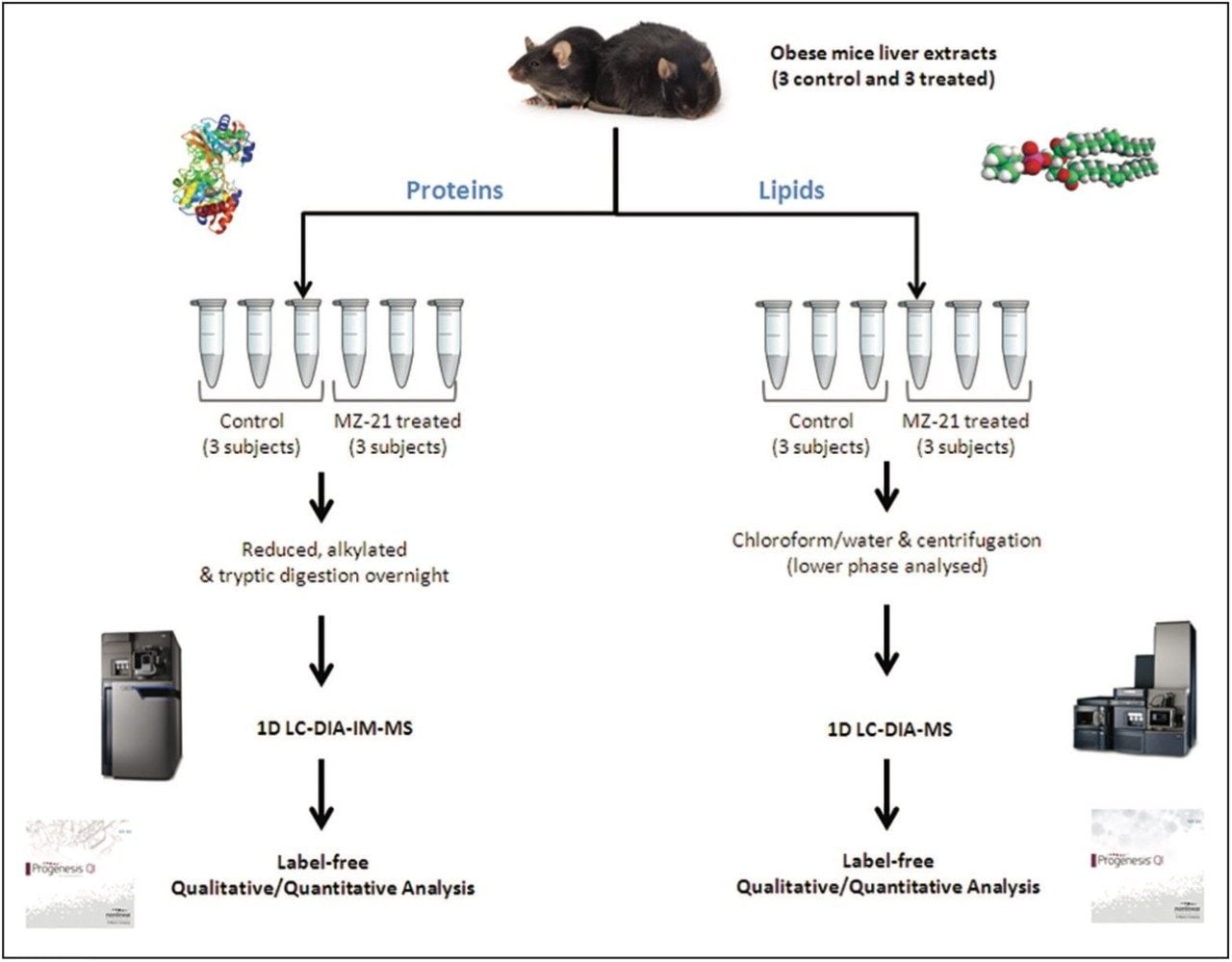
Proteins and lipids were extracted from liver tissue from three control mice and three obese mice treated with MZ-21 glucosylceramide inhibitor. The protein extracts were prepared with 1% RapiGest SF prior to reduction and alkylation and overnight digestion with trypsin.
Lipids were extracted from pre-weighed liver tissue and homogenized in chloroform/methanol (2:1, v/v), in accordance with the Bligh and Dyer method.2 Centrifugation, for 15 min at 4 °C, yielded phase separation, with the lipid-containing (lower) phase isolated for LC-MS analysis.
The LC-MS peptide and lipid data were processed and then searched using Progenesis QI. Progenesis provided normalized, label-free quantification along with peptide and compound identifications. Additional multivariate analysis of the data was performed using EZInfo statistical analysis software.
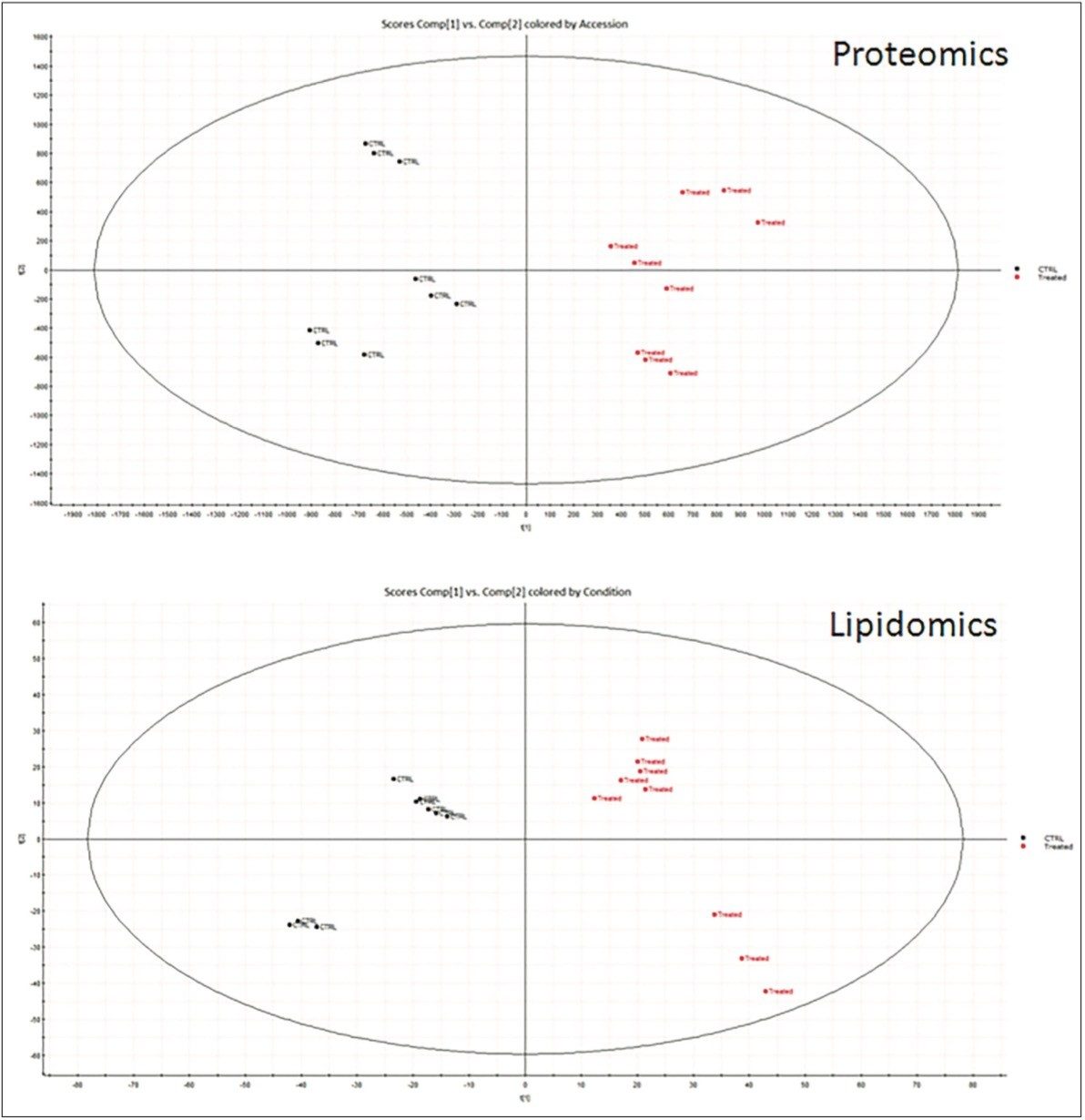
Tissue extracts from the livers of obese mice were analyzed to identify, quantify, and determine protein and lipid variances between the control mice and those treated with MZ-21 inhibitor (Figure 1). Unsupervised, principal-component analysis (PCA) in both data sets clearly shows separation between the control and treated subjects (Figure 2).
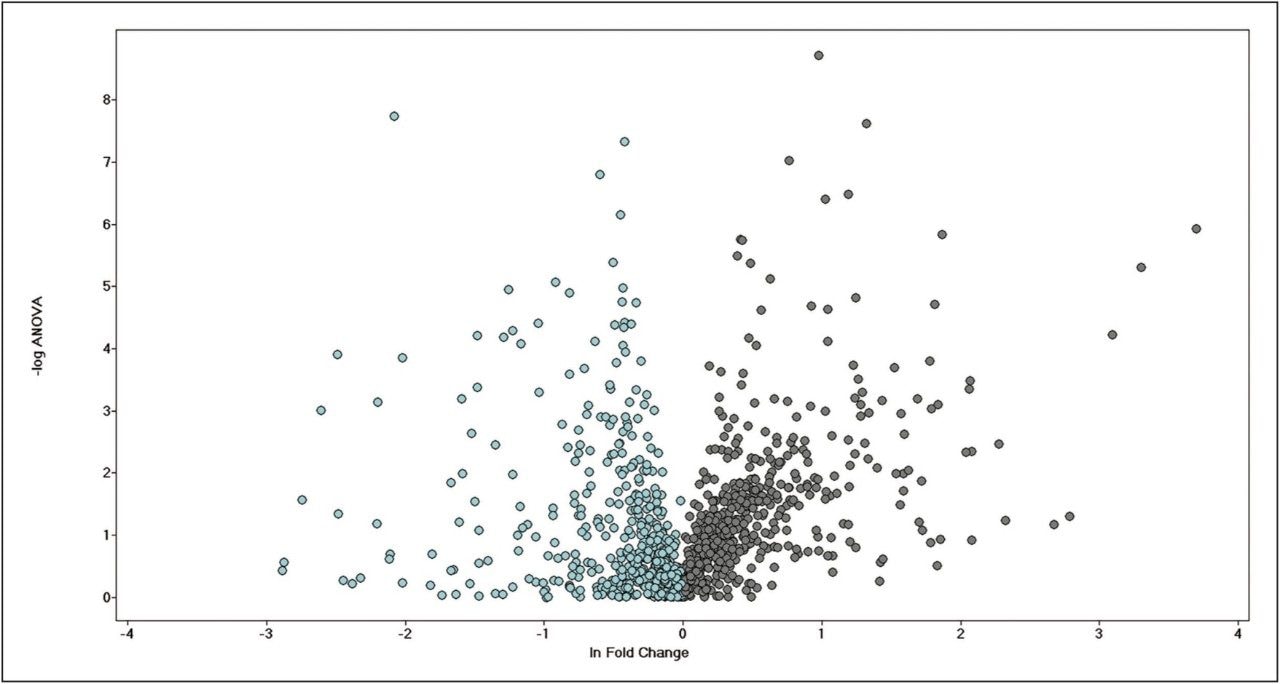
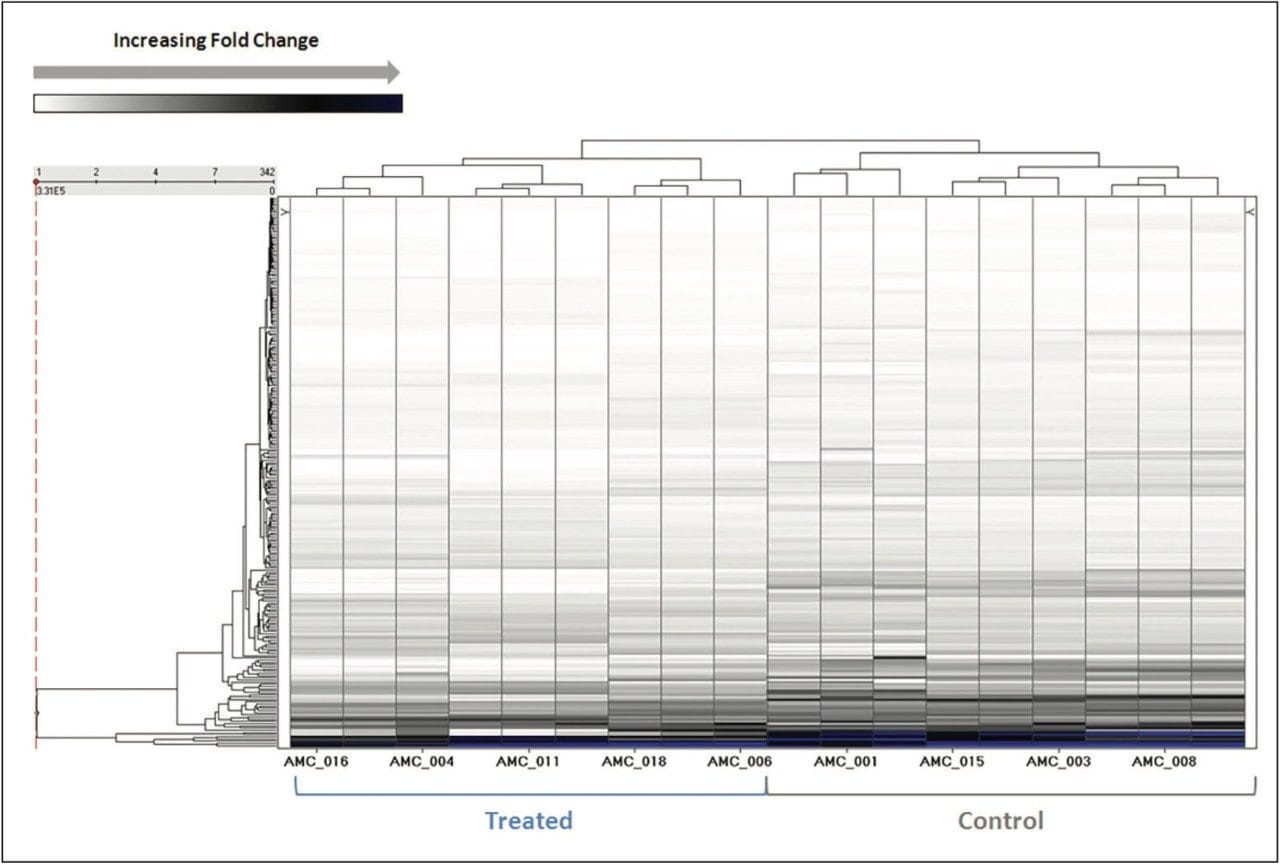
More than 1200 proteins were identified, based on a minimum of 1 peptide per protein and a false discovery rate of 4% (protein level). A volcano plot representing the entire data set readily identifies individual proteins, showing statistically significant ANOVA (p) value changes, indicating protein expression, between the control and treated subjects (Figure 3). Additional filtering of the data ensured that only proteins with a maximum fold change >2 and ANOVA (p) value ≤0.05 were included for further analysis. Figure 4 represents hierarchal clustering of the filtered proteins, with primary grouping at the technical level and secondary grouping at the sample-group level regulation probability.
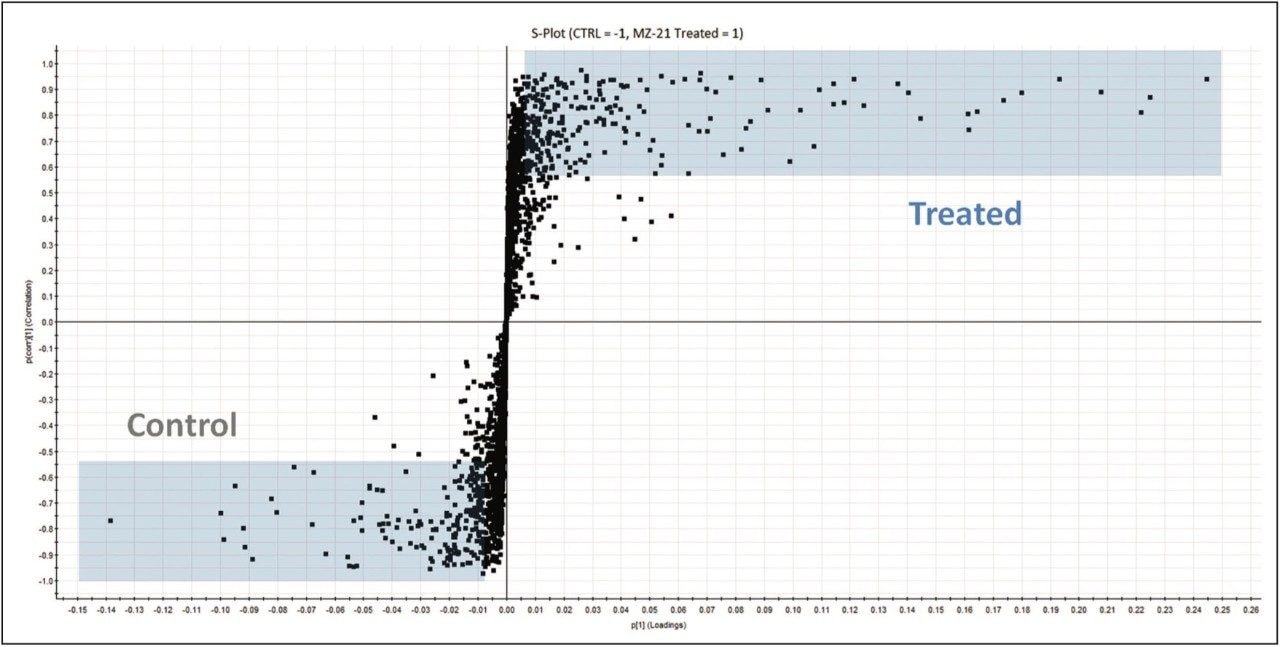
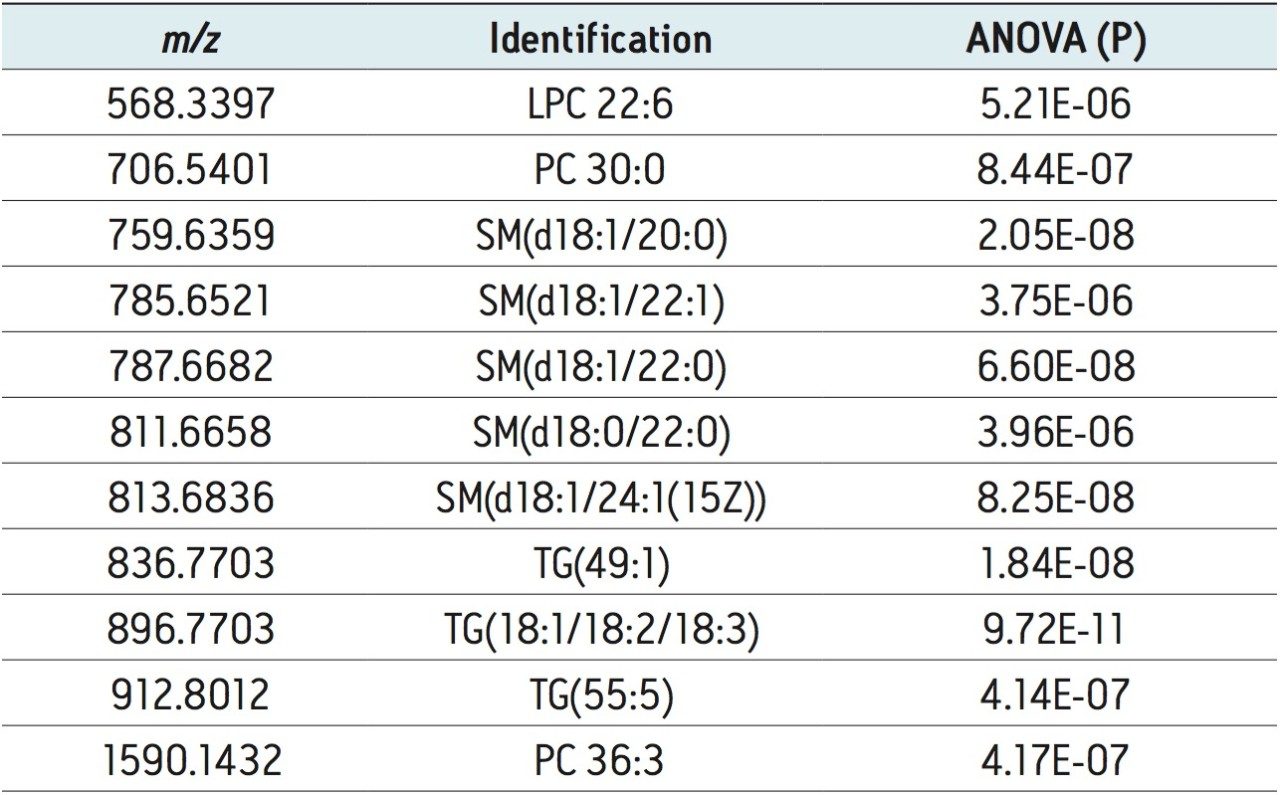
The lipidomic workflow results are summarized in Figure 5. An S-plot constructed from the unsupervised PCA scores displays the metabolites’ contribution to the model (x axis) and the reliability of the measurement (y axis). Thus the plot highlights significant features subject to database searches for tentative lipid identifications. Example identifications providing the most significant variance are shown. They include phosphatidylcholines (PC), sphingomyelins (SM), triglycerides (TG), and lysophosphatidylcholines (LPC). Stringency to the data was achieved by ensuring that only identifications with mass accuracy (<3 ppm), ANOVA (p) value (≤5E-5), and fold change (>2) were retained for further analysis.
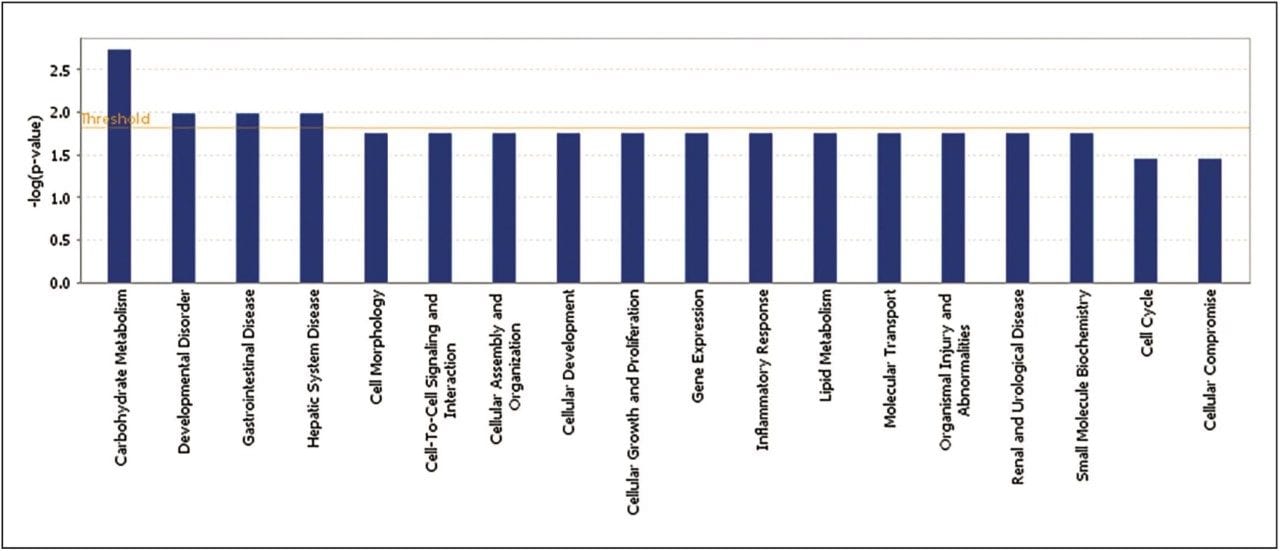
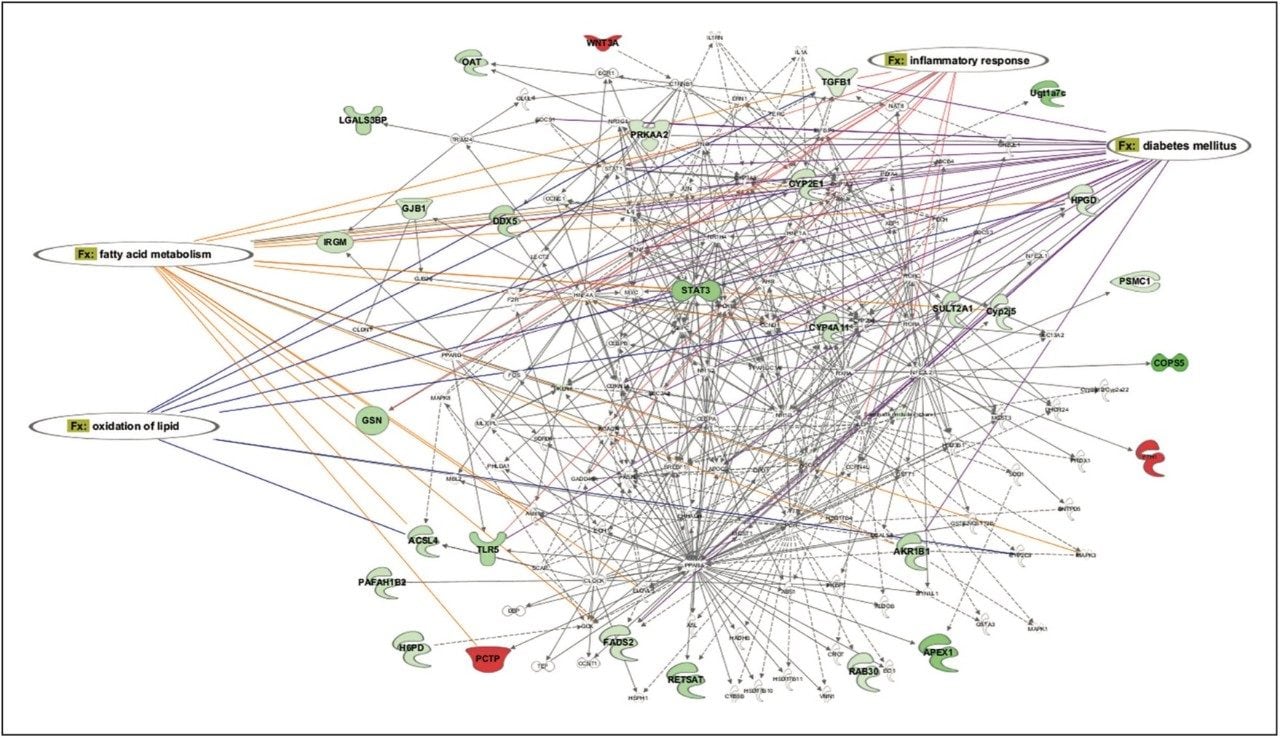
Statistically significant proteins and lipids exhibiting regulation-probability values, identified in both sample groups, were interrogated by means of Ingenuity Pathway Analysis (IPA) software. Various pathways were identified as significant contributors to a range of downstream effects, which include disease and biological processes such as glycogenolysis and inflammatory and gastrointestinal disorders (Figure 6). A core analysis involving both proteomic and lipidomic data, revealed networks involved with lipid metabolism and molecular transport. Diseases commonly associated with obesity, such as diabetes and inflammation, are significantly identified, and they show a number of key components down-regulated following treatment with glucosylceramide inhibitors (Figure 7).
720005092, April 2016