
This is an Application Brief and does not contain a detailed Experimental section.
Waters Metabolite Identification Application Solution provides the flexibility to capture, manage, and share information in a way never before possible - leading to greater scientific understanding of metabolic processes in the discovery and development of new drugs.
The flexibility of the UNIFI Software architecture provides users with the ability to:
In recent years, drug metabolism studies have been facilitated greatly by intelligent informatics designed to deliver answers faster and with more confidence. However, while accessible, these approaches and key informatics value-added tools have not been always been ideally integrated within the true analytical and reporting workflows. The focus has clearly been on developing software tools to answer specific questions, with the consequence of an in-built lack of flexibility to address diverse experimental requirements. Along with this, developers have historically designed software packages to operate independently from the constraints of further analysis and communication of results, although for most scientists, this has become the largest bottleneck in their workflow. For example, if a reinjection is required, a new experiment is initiated and the new data is not linked to the original study (within the software package).
Throughout Waters’ long history of working with the pharmaceutical industry, a consistent thread of dialogue occurs. In order to achieve the best results for their studies, scientists not only need state-of- the-art UPLC and mass spectrometry systems with the ability to capture the most complete datasets, but also powerful software that takes data and converts it to information that forms the basis of decision-making.
Waters Metabolite Identification Application Solution continues this tradition integrating UPLC-MS with data analysis and data management tools. This next-generation platform provides the flexibility to capture, manage, and share information in a way never before possible – leading to greater scientific understanding of metabolic processes in the discovery and development of new drugs.
With UPLC and mass spectrometry techniques such as MSE, MS/MS, and DDA, analysts can extract and enormous amount of information from a sample. The goal of capturing this information is to answer a specific question. With UNIFI, you are free to create and implement your own view of the data, which tools to apply, and in what order, giving you the flexibility to get the answers you seek.
In this way, information can be collated, aligned, identified, annotated, and presented in a customized format so that decisions relevant to your studies can be made quickly and with confidence.
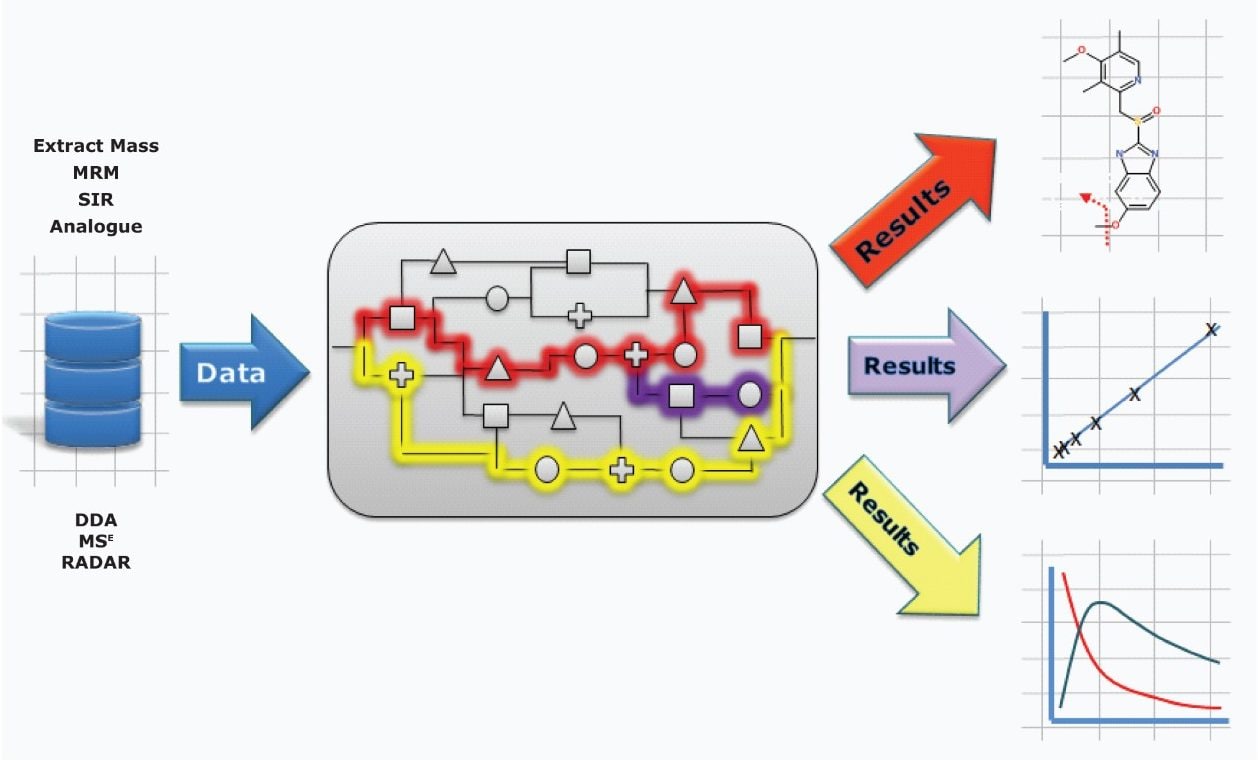
Built upon the industry goal standard workflows of MetaboLynx XS v.2.0 and bringing forward tools already implemented for the Regulated Bioanalysis Platform solution, these comprehensive qualitative and quantitative workflows are now also available for exact mass analyses.
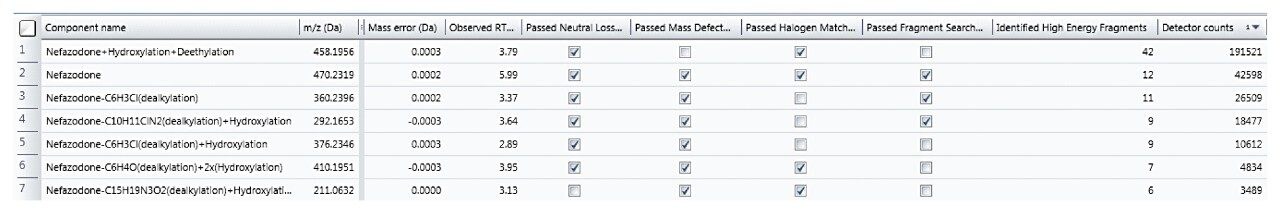
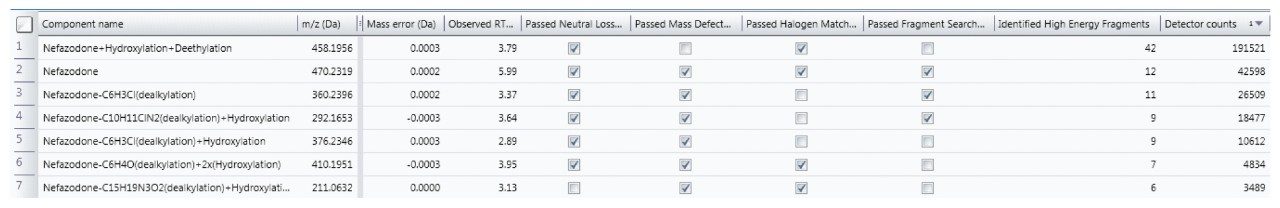
Rather than taking evidence in isolation and eliminating components based upon one piece of evidence (such as MDF), many common characteristics to the parent are now taken into consideration to provide a more balanced view of data and ensure that atypical metabolites are not missed.
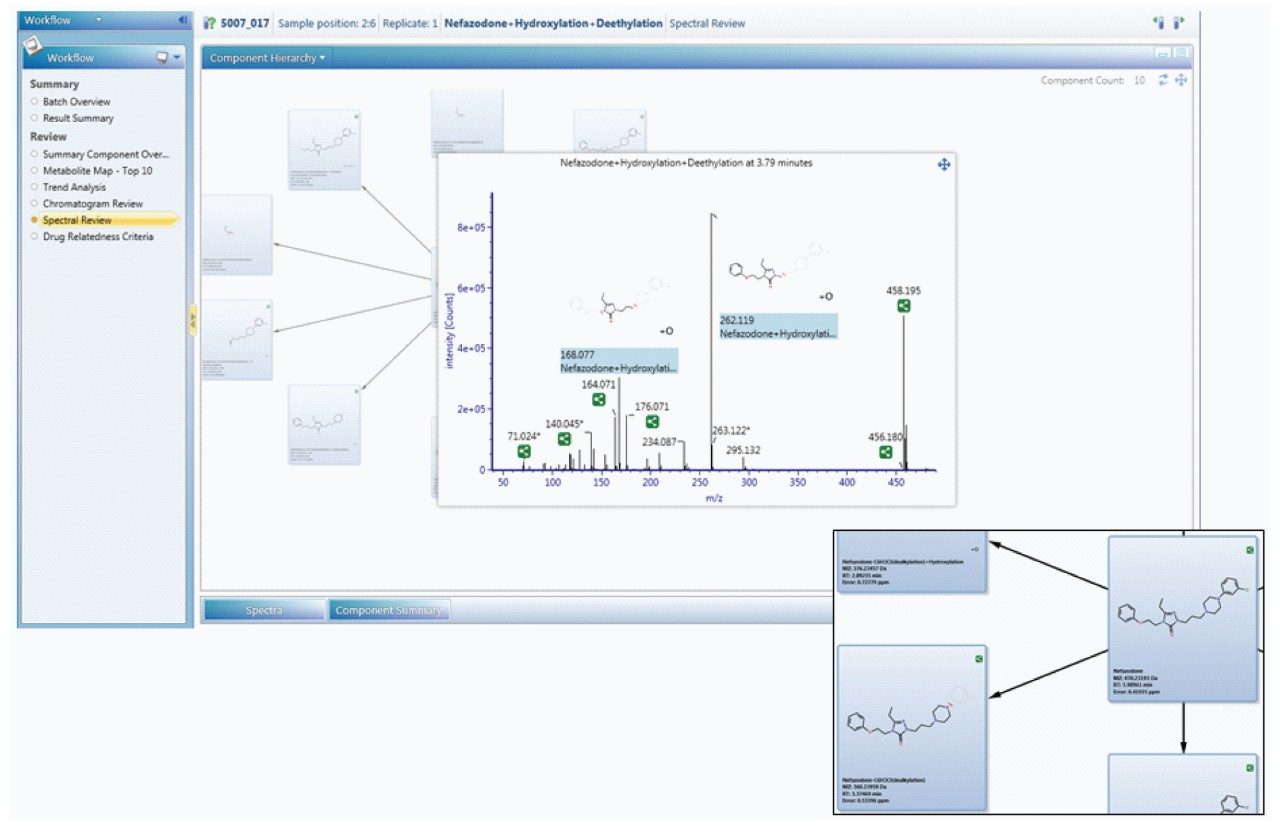
The Drug Related Properties view provides a comprehensive overview of why a component is selected as a potential metabolite.
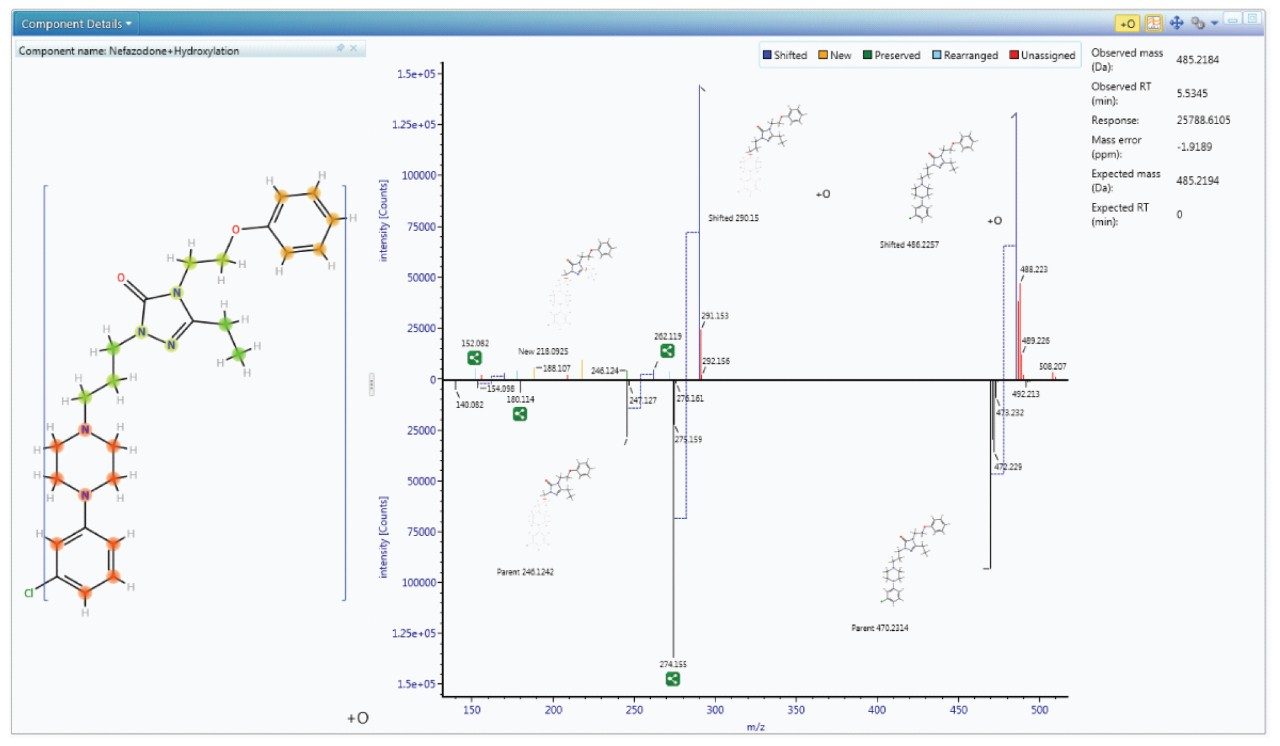
Since metabolism is confidently assigned with MSE product ion spectra are collected for every detectable ion, UNIFI automatically uses this information to assign the site of biotransformation. This is performed for all metabolites that have fragment ion spectra available. The information is annotated both as a heat map and as a bracketed area selection for upmost clarity when sharing this information with colleagues.
Once identified, the metabolism map view allows you to quickly see the most pertinent information, such as the identification, potential structure, precursor and product ion spectra – all in one place, for all metabolites.
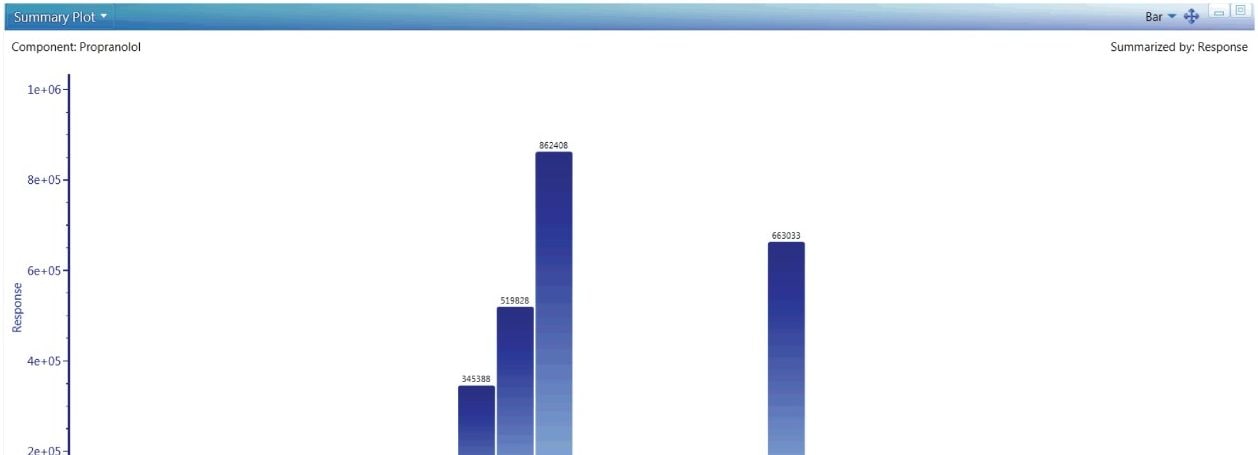
The role of exact mass instrumentation is no longer restricted to characterization. QuanTof detectors deliver more dynamic range at faster speeds than any other technology. UNIFI’s quantitative workflows are designed to take advantage of this technology, giving users the option of easily performing experiments, such as intrinsic clearance, bioavailability and CYP inhibition, and phenotyping. Figure 5 shows the built in tools to track and measure the relative quantitative differences across a sample set.
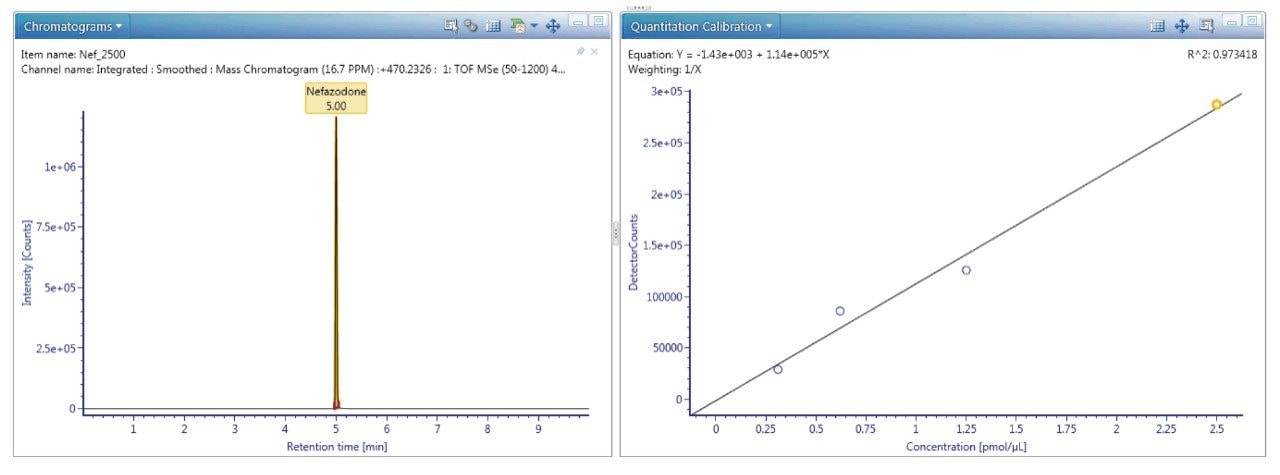
UNIFI’s quantitative tools are built for regulated Bioanalysis laboratories and are routinely available for exact mass use. If more rigorous quantitation is needed, these tools provide upmost confidence in the quality and completeness of the results returned, as shown in Figure 6.
With a database at the core of the software, the UNIFI scientific information system has been built to securely capture and store not only data, but also methods, measurements, structures, identifications, figures, tables – all of the information gathered during the study.
In a workgoup deployment this information is available on-demand and can be accessed anytime during your analyses, bringing knowledge from previous experiences to bear on your work today.
UNIFI’s easy-to-use built-in templates allow users to create fully customized reports that can include any information required, such as logos, descriptions, figures, data, and calculations, as shown in Figure 7. These reports are available for review and sign off as hard copies, and they can always be accessed electronically on a restricted user-based access. If needed, you can output any view for easy integration with existing platforms.
Waters Metabolite Identification Applications Solution with UNIFI redefines the way that users can interact with data collected in drug metabolism studies. By incorporating the appropriate data management tools, analysts can focus on their experimental needs – generating results to drive their science forward. With the added ease of distribution and dissemination of information, a significant barrier to productivity has been removed and future access to the information produced in your organization is guaranteed.
720004323, May 2012