Fast and Sensitive In Vitro Metabolite Identification of Verapamil Using UPLC With Xevo G2 QTof
This is an Application Brief and does not contain a detailed Experimental section.
Abstract
To identify the metabolites of 1 µM verapamil obtained from an in vitro human liver microsome incubation using the ACQUITY UPLC®/Xevo™ G2 QTof with the MetaboLynx™ XS Application Manager Software.
Benefits
Introduction
In recent years, as more high profile drugs are withdrawn from the market due to safety concerns, drug metabolism and toxicity studies have received more attention during the drug R&D process. There is a clear trend towards performing drug metabolism studies earlier in the drug discovery and development stage. A common practice is to conduct an in vitro metabolism study of the parent drug so that the soft spot of the drug can quickly be identified early in the process.
One of the challenges for metabolic identification in the discovery stage is the need for rapid, generic methods, which are sufficiently sensitive such that in vitro incubation studies can be conducted at low µM concentration levels, allowing a closer approximation to how compounds behave in vivo.
A typical in vitro metabolic study includes the investigation of both rate and metabolic routes of the parent drug. A desirable analytical protocol for this type of study needs to provide both the analytical speed and the sensitivity to detect metabolites at substrate concentrations which mimic in vivo conditions.
Results and Discussion
Verapamil was incubated at 37 °C with human liver microsome at 1 µM concentration, with the reaction terminated at 0, 15, 30, 60, 120, and 240 min respectively by adding an equal volume of cold acetonitrile. The samples were centrifuged and the supernatants were injected directly.
Chromatographic separation was performed using Waters® ACQUITY UPLC with an ACQUITY UPLC HSS T3, 1.7 µm, 2.1 x 100 mm Column. The mobile phase consisted of water with 0.1% formic acid (A) and acetonitrile (B). The injection volume was 5.0 µL. The Xevo G2 QTof Mass Spectrometer was used in positive ESI mode for data acquisition using UPLC/MSE , which allows both precursor and product ion data to be acquired in one injection.
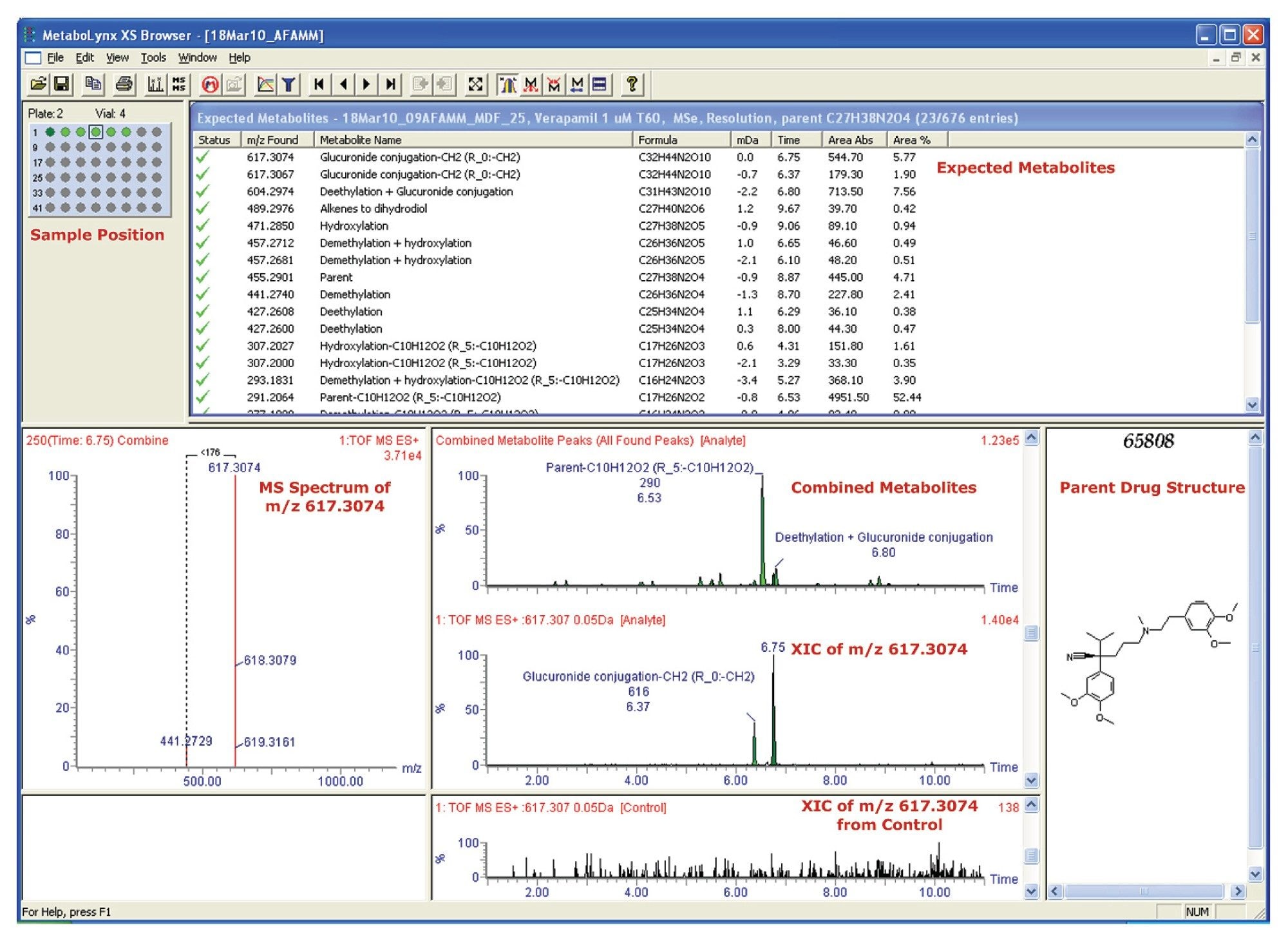
MetaboLynx MS was used for data mining and results were displayed in the MetaboLynx Browser, as shown in Figure 1.
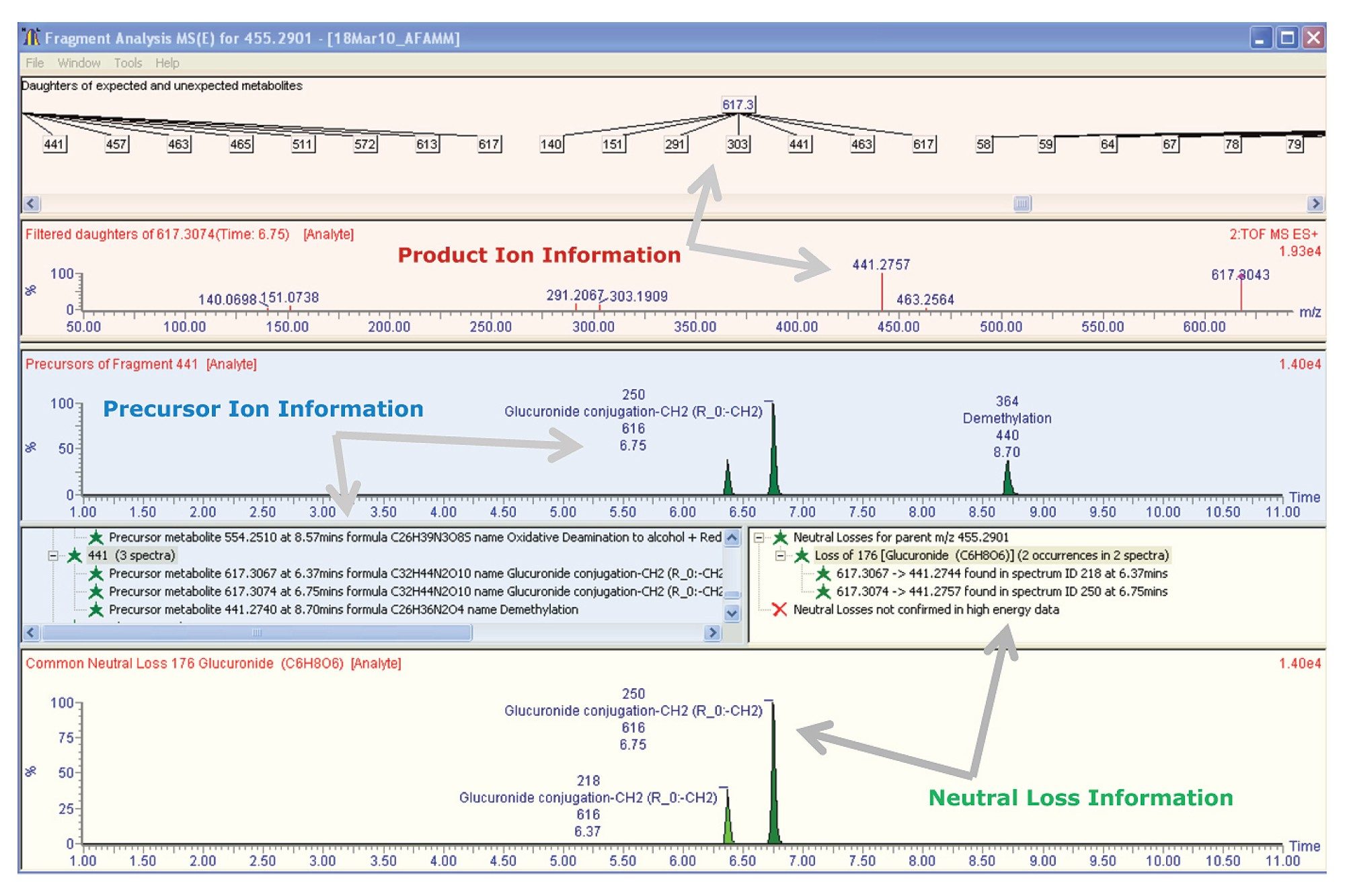
Product ion information was processed at the same time and displayed in the Fragment Analysis Window within the MetaboLynx Browser (Figure 2).
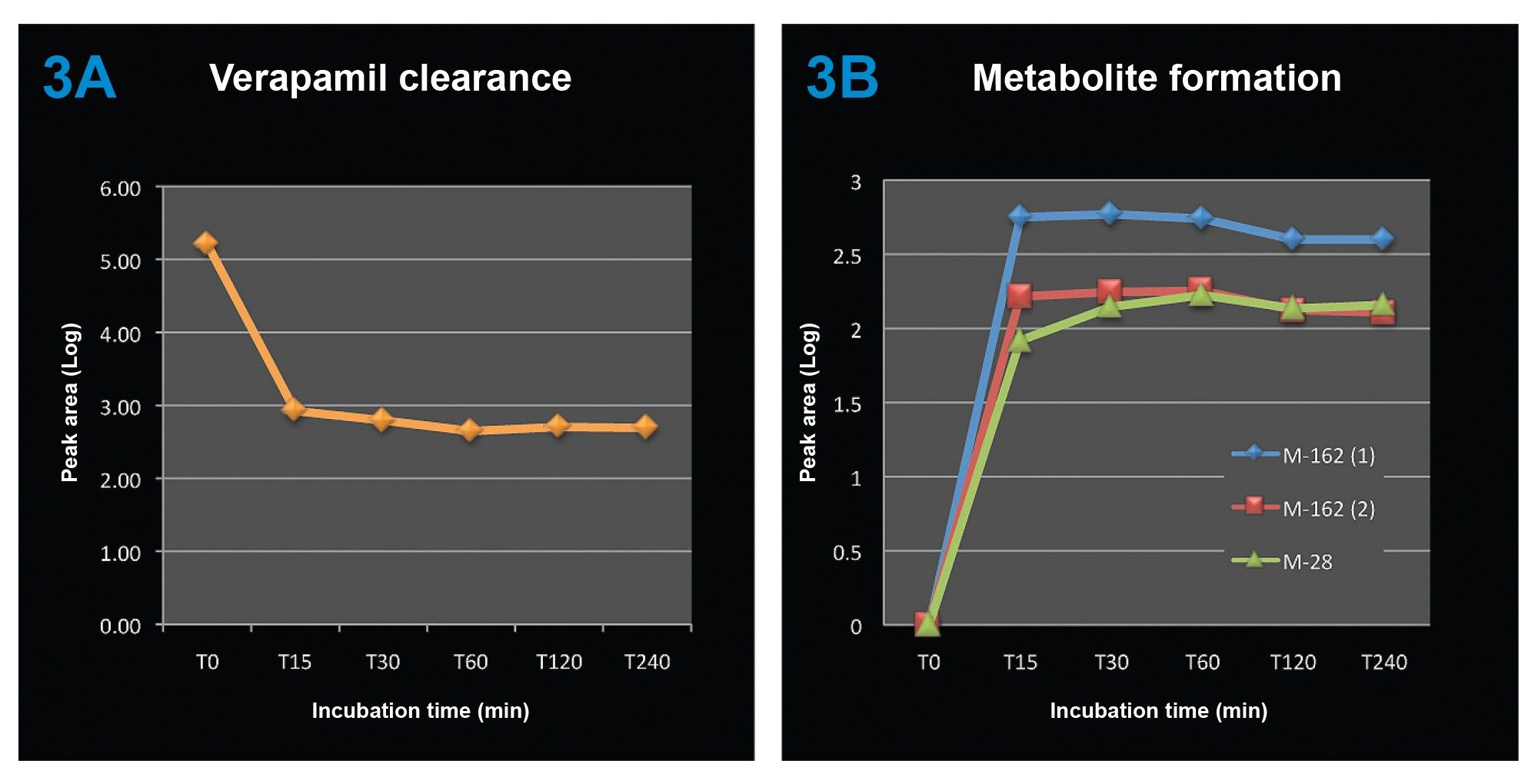
With multiple time points injected, the parent drug clearance curve and the metabolite formation curves were obtained during the same experimental run, as shown in Figure 3.
Conclusion
This technical brief demonstrates that by utilizing the Xevo G2 QTof with UPLC/MSE and MetaboLynx XS workflow, an in vitro metabolite study can be performed at low µM levels with speed, sensitivity, and selectivity.
The Xevo G2 QTof, with its innovative QuanTof™ Technology and Engineered Simplicity,™ ideally integrates with an ACQUITY UPLC System. It is the most sensitive benchtop oaTOF instrument available, has a 1 ppm exact mass measurement capability, and data acquisition speeds of up to 20 spectra/sec.
By using a UPLC/MSE data acquisition strategy coupled with the MetaboLynx XS data processing workflow with chemical intelligence, the complete metabolite identification task can be accomplished rapidly from a single LC injection. With multiple time-point samples injected, the rate and routes of metabolism of a target drug can be easily obtained at low µM incubation levels. As a result, the goal of maximizing productivity is easily obtained.
Featured Products
720003425, May 2010