For research use only. Not for use in diagnostic procedures.
This is an Application Brief and does not contain a detailed Experimental section.
In this technology brief desorption electrospray ionization (DESI) mass spectrometry imaging (MSI) was used to analyse tissue sections from cassette dosed drug metabolism and pharmacokinetic (DMPK) models. The ability to map the distribution of drugs and their metabolites in the liver, alongside endogenous lipid species is demonstrated.
Mass spectrometry imaging for the distribution of drugs and their associated metabolites, a DMPK solution.
Assessing the ability of drug candidates to reach the desired target(s) in preclinical animal models is an essential step of the pharmaceutical discovery and development process. Typically, whole-body autoradiography (WBA) is the method routinely used for assessing pharmacokinetic and distribution properties of a drug candidate.
This technique requires the introduction of a radiolabel to the synthesis of the drug, and then allows the visualization of the radiolabel in a quantitative, sensitive manner. However, as it relies on the detection of the radiolabel, WBA is unable to distinguish between the parent compound and any potential metabolites that could also contain the radiolabel. Furthermore, to synthesize the radiolabeled compound often requires an alteration in the chemistry used, and can be an expensive procedure.
Mass spectrometry imaging (MSI) permits the visualization of molecules in a label-free and multiplexed approach. This provides the accurate localization of drug candidates and potential metabolites simultaneously, without the need for labeling. There is also potential for a better understanding of the mechanism of action of drug candidates, and the ability to identify m/z values for use as biomarkers and companion diagnostics. Multimodal MSI approaches are known to give complementary information.¹ For example, secondary ion mass spectrometry (SIMS) can be used to generate high spatial resolution images (nanometer level) with low compound coverage, and matrix assisted laser desorption ionization (MALDI) can be used for better compound coverage with micrometer spatial resolution. Recently, electrospray (ESI) type ionization techniques, such as desorption electrospray ionization (DESI) and liquid extraction surface analysis (LESA), have been implemented for MSI experiments.
In this experiment using an orally dosed mouse model, one cohort of animals was cassette dosed with four drugs (Terfenadine, Olanzapine, Moxifloxacin, and Erlotinib, at 25, 10, 10 and 25 mg/kg respectively) and a second cohort was only dosed with Olanzapine at 25 mg/kg. A further cohort was vehicle dosed to act as a control. Animals from each cohort were euthanized at either 2 or 6 hrs post administration and liver sections were then taken.
DESI-MSI analysis was performed using a Prosolia 2D DESI source with a Waters sprayer on a Waters Xevo G2-XS QTof Mass Spectrometer with data acquisition and processing through Waters High Definition Imaging (HDI) v1.4 Software. Ion images demonstrating the localization of the four drugs as well as endogenous species (mainly lipid species) were generated from a single imaging experiment, as shown in Figure 1. The distribution and signal intensity of ions across the sections were consistent with those shown in previously published work.² In all figures, where no ions were detected, dotted outlines of the tissues are shown for reference. All data were acquired using a step size of 150 µm and subsequently normalized.

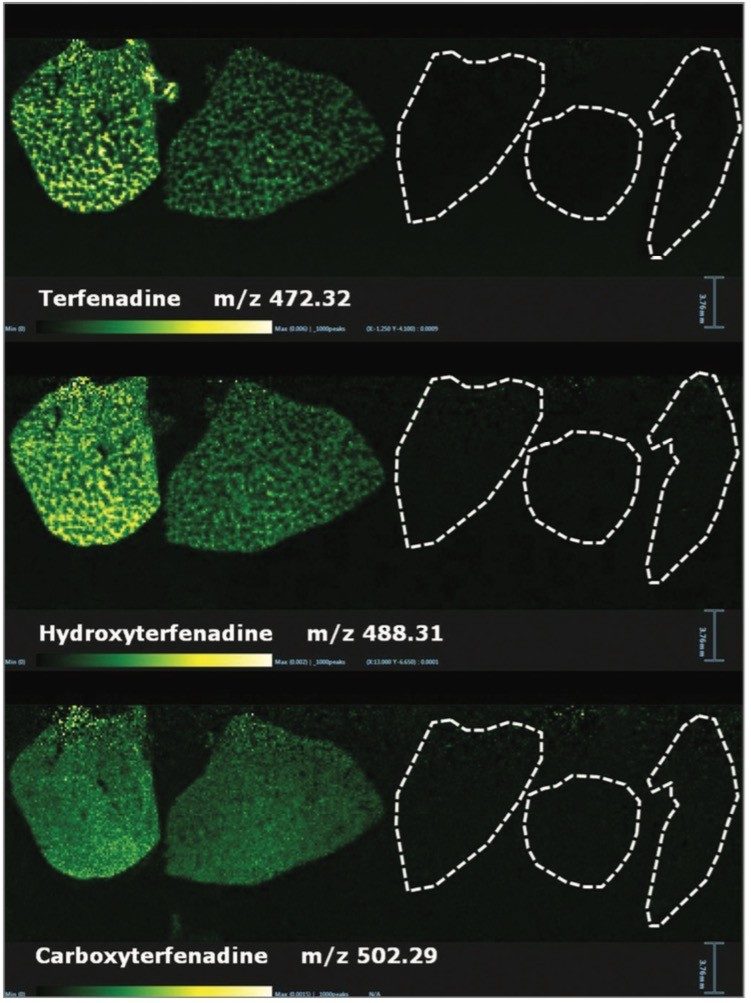
Within the same experiment it was also possible to detect some of the metabolites from Olanzapine, Terfenadine, and Erlotinib. The ion images for Terfenadine (m/z 472.32) and for two of its metabolites: hydroxyterfenadine (m/z 488.31) and carboxyterfenadine (m/z 502.29) are displayed in Figure 2.
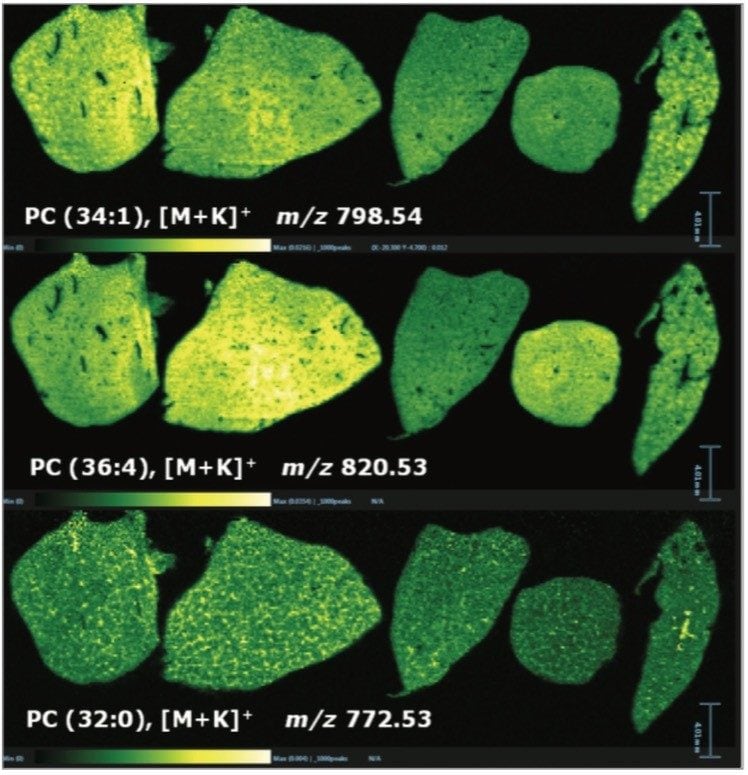
In addition to the information gained for the distribution of the drugs themselves and their metabolites, it is also possible to view the distribution of molecular ions from endogenous species. Figure 3 shows the varying distribution of endogenous phosphatidylcholine lipid species (as labeled on the individual images).
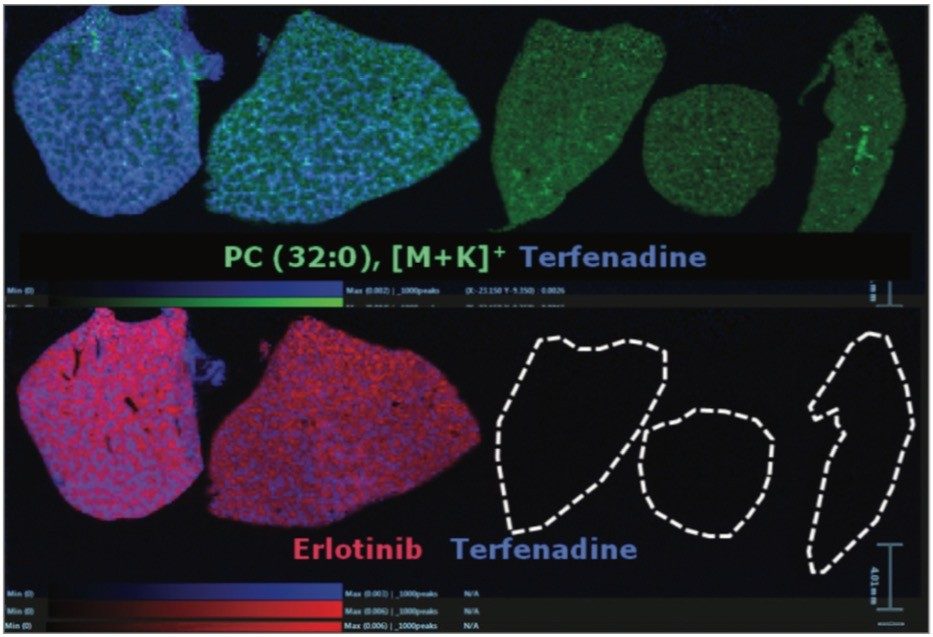
Overlaying the drug, metabolite and lipid ion images, Figure 4 shows RGB overlaid ion images, allowing clear visualization of the different spatial distribution of ions within different anatomical microstructures in the liver. This has the potential to help determine the biological and histological impact of the drugs and metabolites throughout the tissue, and provides an extra level of detail not available with other techniques.
The specific localization of molecules and degree of information obtained can be improved by the acquisition of additional imaging datasets at higher spatial resolution, which, in the case of DESI-MSI is possible using either a consecutive tissue section, or if using solvents of a low abrasive nature, on the same tissue section, as shown in the 150 µm images pictured here.
We thank Dr Richard J.A. Goodwin and John G. Swales from the Drug Safety & Metabolism group at AstraZeneca R&D for providing all the samples used in this technical brief.
720005947, June 2019