For research use only. Not for use in diagnostic procedures.
This application note demonstrates a high-throughput UPLC-MS/MS research method for the semi-quantitative analysis of various tryptic peptides in human serum samples.
Proteins are important molecules that are involved in almost all biological processes. They are large, high molecular weight molecules, and therefore are analyzed using marker peptides that are produced using proteolytic enzymes like trypsin. Historically these types of analyses have been performed using high-resolution mass spectrometry coupled with micro/nano flow chromatographic systems. These methodologies however are low throughput, and are not suitable for large cohorts of samples. Here we demonstrate a high-throughput UPLC-MS/MS research method for the semi-quantitative analysis of various tryptic peptides in non-depleted, tryptically digested human serum samples. This application note is partof a Targeted Omics Method Package.
Human serum samples were prepared using the Biognosys Plasma Dive Kit (Biognosys, Schlieren, Switzerland). Briefly, 10 μL ofsample was denatured, reduced, and alkylated before being diluted and typtically digested using 5 μL of 0.4 μg/μL typsin. Following acidification, centrifugation, and the addition of a fixed amount of the stable labeled forms of all 100 marker peptides, 6 μL of the spiked supernatant was then injected onto the UPLC-MS/MS system.
UPLC separation was performed with an ACQUITY UPLC I-Class System (fixed loop), equipped with a CORTECS T3 2.7 μm (2.1 × 30 mm) analytical column. A sample of 6 μL was injected at a flow rate of 0.15 mL/min. Mobile phase A was 0.01% formic acid (aq) containing 0.2 mM Ammonium Formate and mobile phase B was 50% isopropanol in acetonitrile containing 0.01% formic acid and 0.2 mM Ammonium Formate. After an initial 2.5-minute hold at 1% Mobile phase B, the tryptic peptides were eluted from the column and separated with a gradient of 1–45% Mobile phase B over 2.9 minutes, followed by a 2.5-minute column wash at 85% Mobile phase B. The column was then re-equilibrated to initial conditions. The analytical column temperature was maintained at 60 °C.
Multiple Reaction Monitoring (MRM) analyses were performed using a Xevo TQ-S micro tandem quadrupole Mass Spectrometer. All experiments were performed in positive electrospray ionization (ESI+) mode. The ion source temperature and capillary voltage were kept constant and set to 150 °C and 2.0 kV respectively. The cone gas flow rate was 50 L/hr and desolvation temperature was 650 °C. Cone voltages and collision energies used were those calculated by the Skyline software(MacCoss Lab, University of Washington).
Method information was imported onto the LC-MS system using the Quanpedia functionality within MassLynx. This extend able and searchable database produces LC and MS methods as well as processing methods for use in TargetLynx for compound quantification. Skyline was used for the production of MS methods and data visualization.
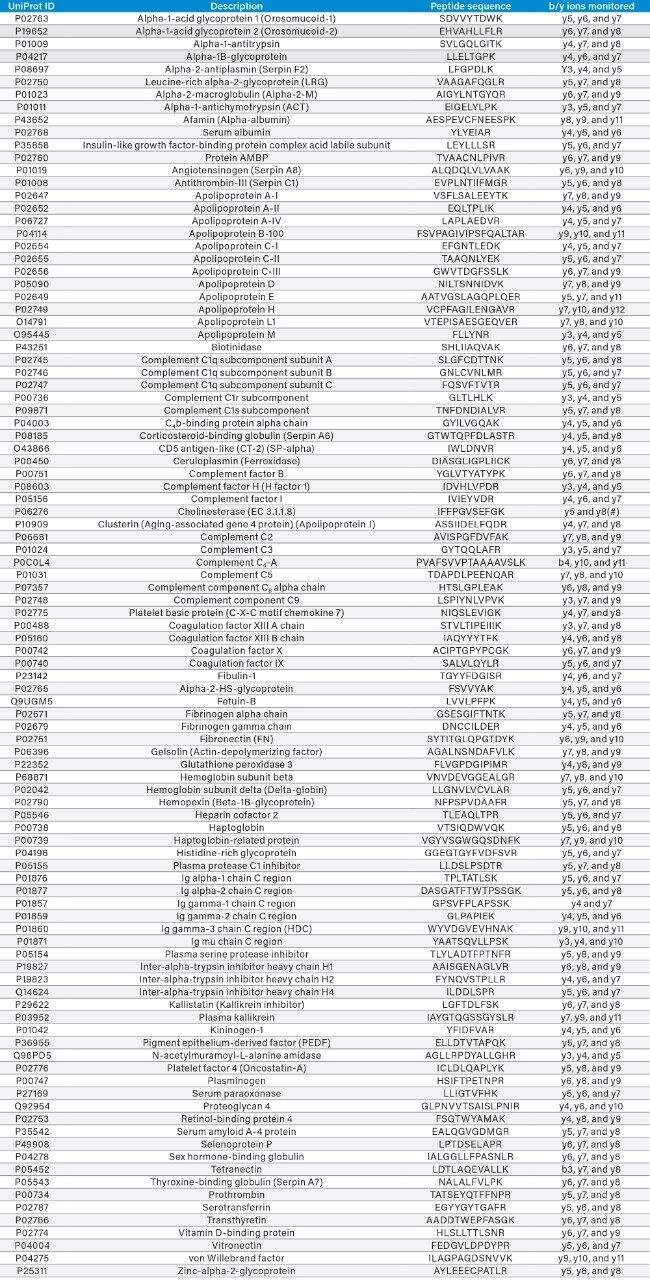
Table 1 details the 100 marker peptides analyzed, the proteins they represent, and the b and y product ions monitored. Tryptic peptides were detected using a series of MRM transitions. The product ions monitored are detailed in Table 1. These were all singly charged ions, with the exception of the y8 ion for P06276, where both singly and doubly charged ions were monitored. The precursor ions used were the doubly charged ions for all marker peptides, with the exception of P08603 and Q9PD5,where the triply charged precursors were used.
# - Both singly and doubly charged ions monitored for.
Table 1. Names, Uniprot ID’s, and marker peptides used for the 100 proteins monitored using the Biognosys Plasma Dive kit. Column 4 details the b and y product ions monitored for the given marker peptide.
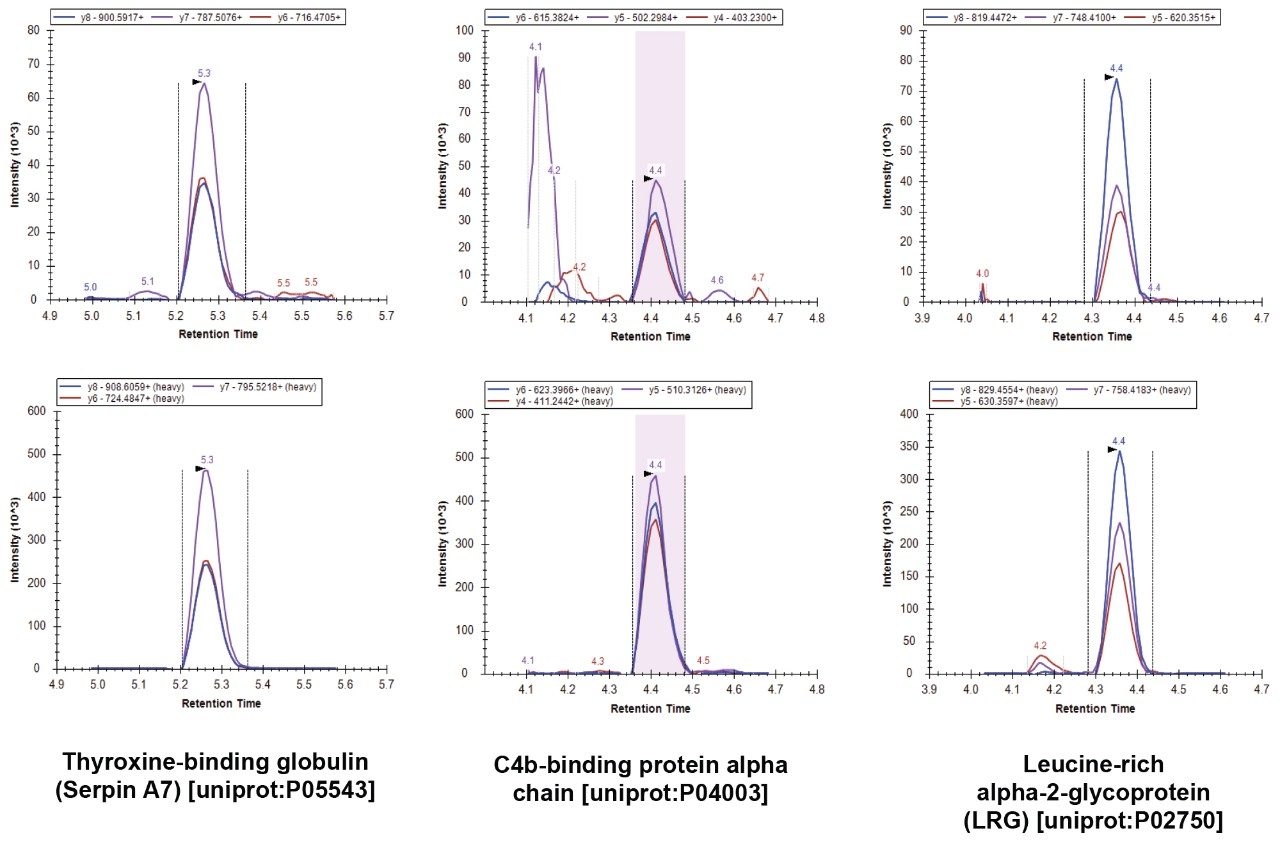
Figures 1 and 2 show example data acquired for six of the 100 proteins. Data for these 100 proteins was acquired over two analyses. Each 12-minute analysis was capable of analyzing 50 marker peptides, where three transitions were monitored for both the native and stable labeled forms. If fewer transitions per peptide were monitored, it would be possible to analyze more proteins in a single injection. However, monitoring three transitions per peptide increased the confidence in identification.
A rapid UPLC-MS/MS methodology has been developed for the research analysis of proteins. This method has been demonstrated to be suitable for the analysis of physiologically relevant levels of multiple proteins inhuman serum. This method utilizes a generic LC-MS platform that can be used for various compound classes (including metabolomics, lipidomics,and proteomics). Deployment of this method in conjunction with other complementary methods available on the Waters MetaboQuan website a can canform the basis of a comprehensive suite of targeted multi-omic workflows.
720006323, August 2018