This work demonstrates the modernization of the USP monograph for high-throughput identity test of insulin analogues.
The fast growing market of biotherapeutics brings certain challenges for pharmaceutical development and quality control, such as throughput, robustness, and cost. This in part is due to using the United States Pharmacopeia USP monographs that were developed using dated instruments and methods. While USP monographs offer the distinct advantage of not requiring re-validation when deployed in the routine analysis of biotherapeutics, these analyses often require excessive resources, such as long analysis time and large solvent consumption. As of 2011, the USP has embarked on an initiative to modernize more than 2000 monographs, including both physiochemical tests and bioassays, deviation from which will require justification on the manufacturer’s part. As part of a sound pharmaceutical system, the International Committee on Harmonization (ICH) recommends continual improvement, including the evaluation of new and innovative technologies and methods to ensure product quality and safety.1 To this end, method modernization affords pharmaceutical companies the ability to address challenges associated with manufacturing for rapidly growing markets with efficient workflows that are aligned with regulatory guidelines while maintaining product quality and safety.
The objective of this work is to demonstrate how the modernization of a USP monograph can increase throughput and productivity in a routine environment. In this study, insulin is selected as an example for its historical precedence and significant market size.2 A new method for identity test will be developed and performed on three insulin analogues to compare with the methods in supporting monographs.
Insulin human, insulin lispro, and insulin glargine were purchased from USP. Endoproteinase Glu-C from S. aureus was purchased from Promega. Digestion and separation procedures were as outlined in the USP monograph: insulin human,3 insulin lispro,4 and insulin glargine.5 New separation methods were also developed as shown in the gradient table. HPLC grade water, acetonitrile, and TFA were purchased from Fisher Scientific and used as received.
|
USP method and scaled USP method |
|
|---|---|
|
LC system: |
Alliance HPLC |
|
Detectors: |
e2489 UV detector, 5 mm flow cell, λ = 215 nm |
|
LC column: |
XSelect CSH C18 Column 3.5 μm, 130 Å, 4.6 mm x 100 mm (P/N 186005269) |
|
Column temp.: |
40 °C for insulin human and lispro 35 °C for insulin glargine |
|
Sample vial: |
12 × 32 mm glass vial, total recovery (P/N 600000750cv) |
|
Mobile phase: |
Water and acetonitrile |
|
MP additive: |
Sulfate buffer for insulin human and lispro, phosphate buffer for insulin glargine |
|
Mass load: |
8.6 μg |
|
LC system: |
ACQUITY UPLC H-Class Bio |
|
Detectors: |
ACQUITY TUV Detector, 5 mm flow cell, λ = 215 nm |
|
LC column: |
ACQUITY UPLC CSH C18 1.7 μm, 130 Å, 2.1 mm × 100 mm (P/N 186006937) |
|
Column temp.: |
40 °C |
|
Sample vial: |
12 × 32 mm glass vial, total recovery (P/N 600000750cv) |
|
Mobile phases: |
Water and acetonitrile |
|
MP additive: |
0.1% TFA |
|
Mass load: |
0.86 μg |
|
Time (min) |
Flow rate (min) |
%A |
%B |
|---|---|---|---|
|
Initial |
0.428 |
90.0 |
10.0 |
|
29.14 |
0.428 |
30.0 |
70.0 |
|
31.57 |
0.428 |
0.0 |
100.0 |
|
34.00 |
0.428 |
0.0 |
100.0 |
|
34.49 |
0.428 |
90.0 |
10.0 |
|
41.77 |
0.428 |
90.0 |
10.0 |
|
Time (min) |
Flow rate (min) |
%A |
%B |
|---|---|---|---|
|
Initial |
0.300 |
85.0 |
15.0 |
|
10.00 |
0.300 |
55.0 |
45.0 |
|
11.00 |
0.300 |
0.0 |
100.0 |
|
13.00 |
0.300 |
0.0 |
100.0 |
|
13.50 |
0.300 |
85.0 |
15.0 |
|
20.00 |
0.300 |
85.0 |
15.0 |
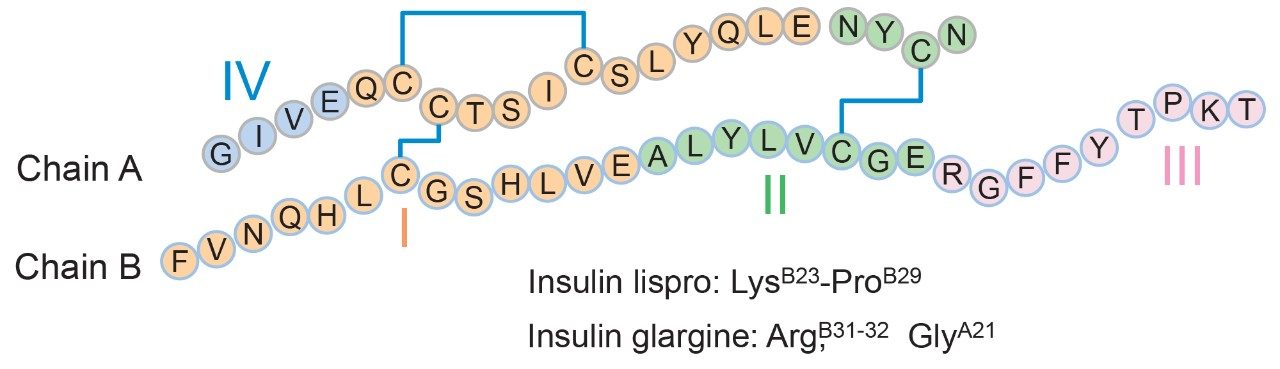
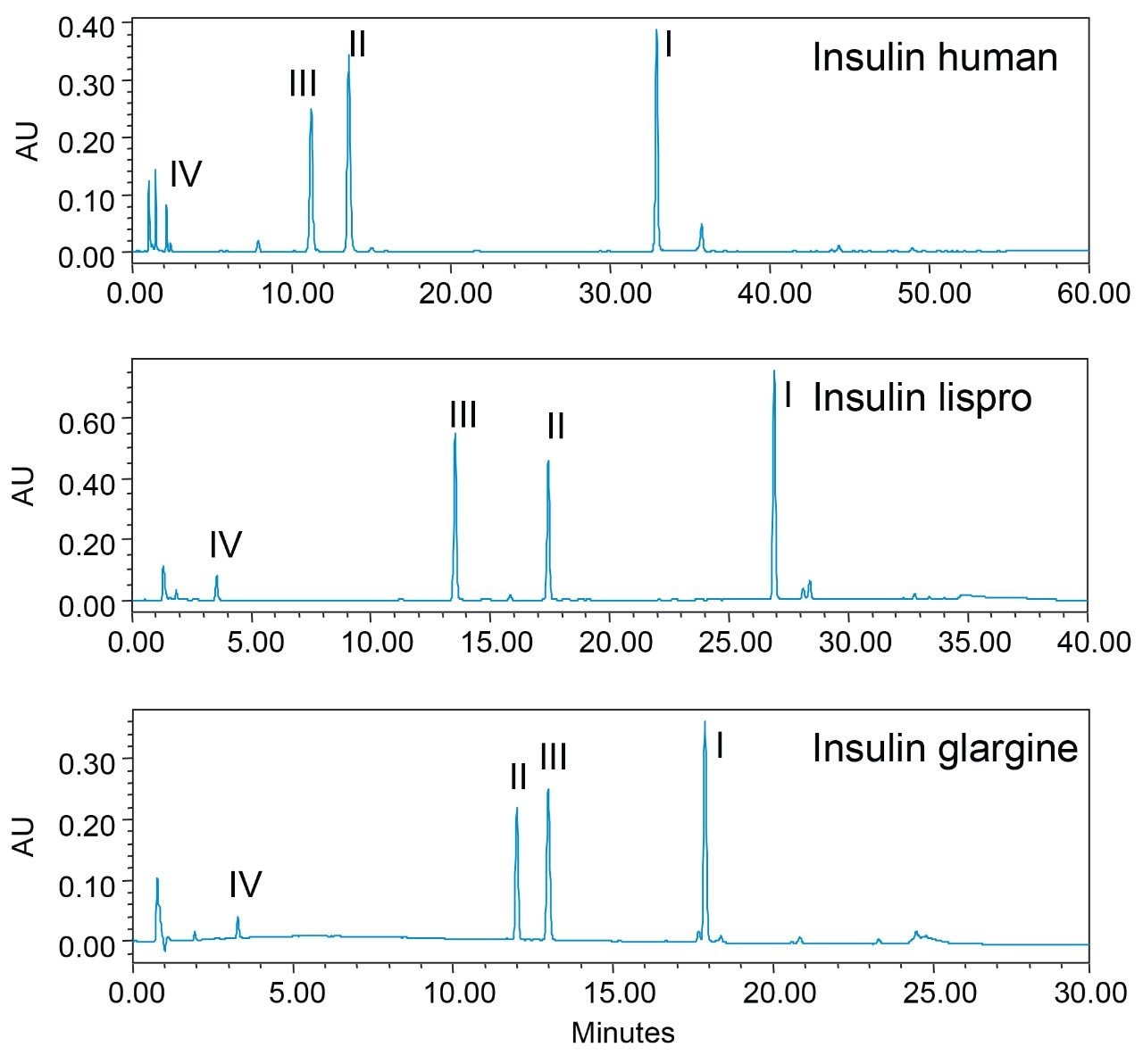
Insulin human is a 5.8 kDa protein that consists of two polypeptide chains (Figure 1). Since its first clinical application in 1922, insulin drugs have undergone rapid development with six analogues now available on the market for different drug activities.6 These analogues have highly similar structures with only slight amino acids difference in sequence. Supporting USP monographs for identity test of each of these analogues also vary from one another with changes in mobile phase, gradient, flow rate, and temperature.3,4,5 To establish a baseline for modernization, the identity tests for these insulin analogues (insulin human, insulin lispro, and insulin glargine) were performed using methods as outlined in their respective USP monograph. An XBridge CSH C18 Column with L1 packing was selected for better peak shape in HPLC separation. Proteolytic cleavage of each analogue produced four peptide fragments that were well resolved using the HPLC method described in each monograph (Figure 2). While sufficient in their current format, it should be noted that the long run times (86 min for insulin human) associated with HPLC can be reduced by updating to a UPLC-based method for improved throughput and reduced waste stream.
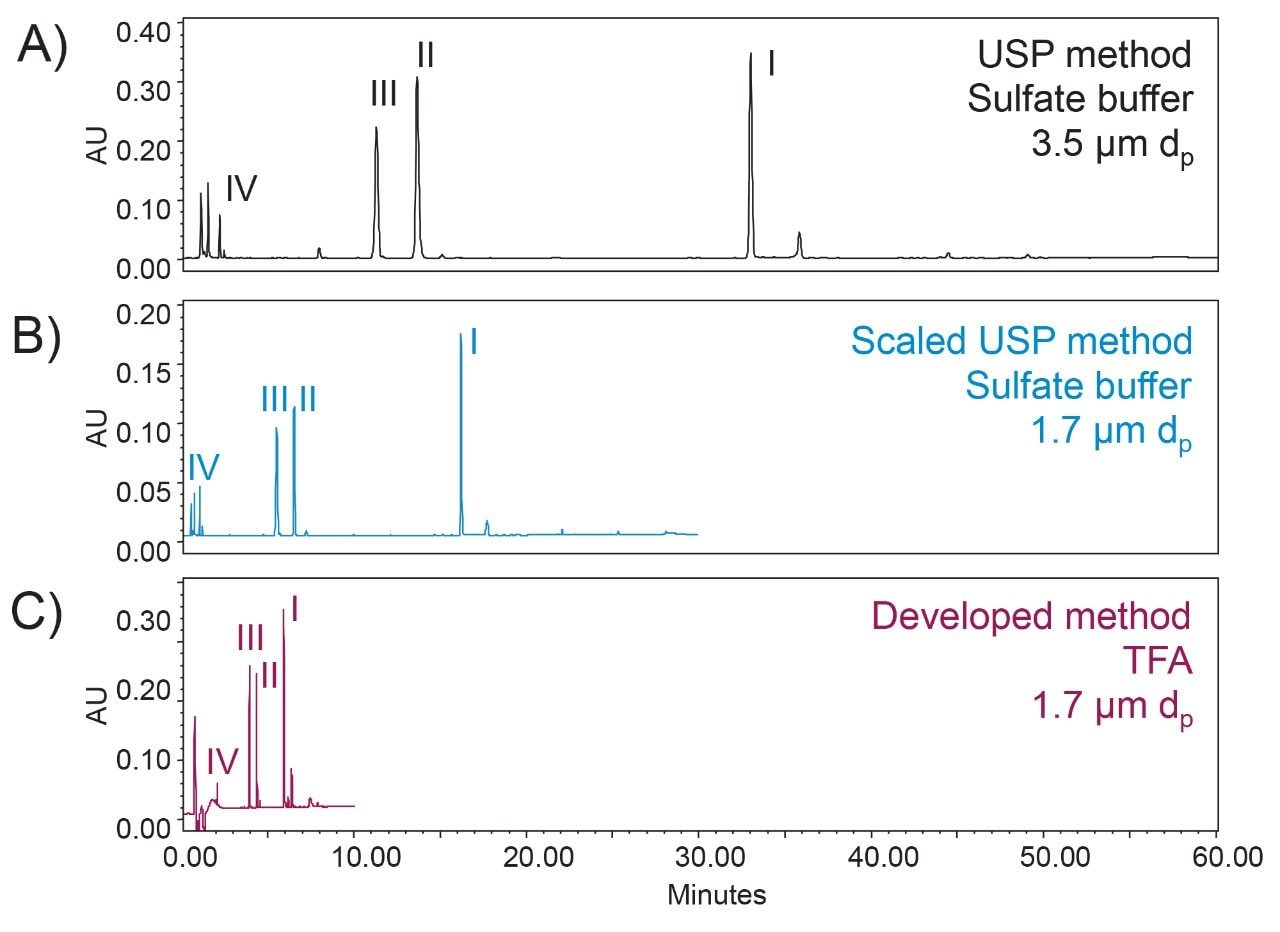
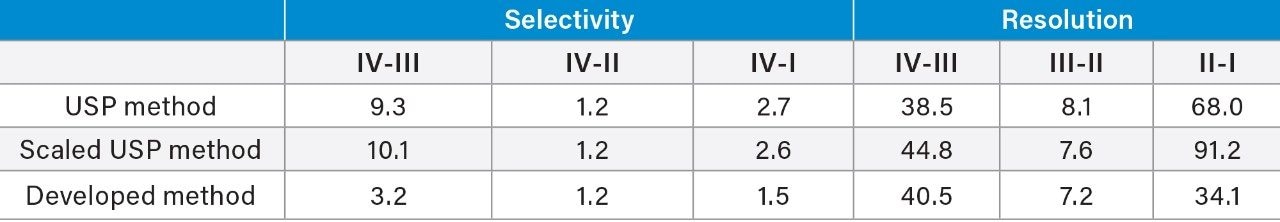
To demonstrate the benefit of a UPLC-based method, USP Chapter <621>7 was referenced for acceptable method changes. As described in <621>, a column with 50% smaller particle size was used along with geometrically scaled gradient conditions to reduce the analysis time. An ACQUITY UPLC H-Class Bio System was used to accommodate the higher pressure generated by columns packed with smaller particles. With a 1.7 µm particle size column, the analysis time was reduced from 60 min (Figure 3A) to 30 min (Figure 3B) for insulin human using the scaled method for improved productivity. It should be noted that the monograph results shown in Figure 2 use different buffer additives which may not be ideal for streamlining the manufacturing process, specifically in the case of a manufacturer with multiple insulin analogues in their product pipeline. To address this challenge we need to consider method development changes beyond USP <621> limitations. With this in mind, TFA was used as a mobile phase additive for the identity test of insulin human. With a 10 min gradient from 15% to 45% mobile phase B, separation time was reduced by 75% from the original USP method as shown in Figure 3C. The impact of TFA on selectivity and resolution was evaluated as shown in Table 1. Comparable selectivity and baseline resolution were observed for the majority of the peptide fragment peaks using the new high-throughput method when compared to the monograph-based methods, suggesting the method could be utilized as a ‘platform’ assay for various insulin analogues.
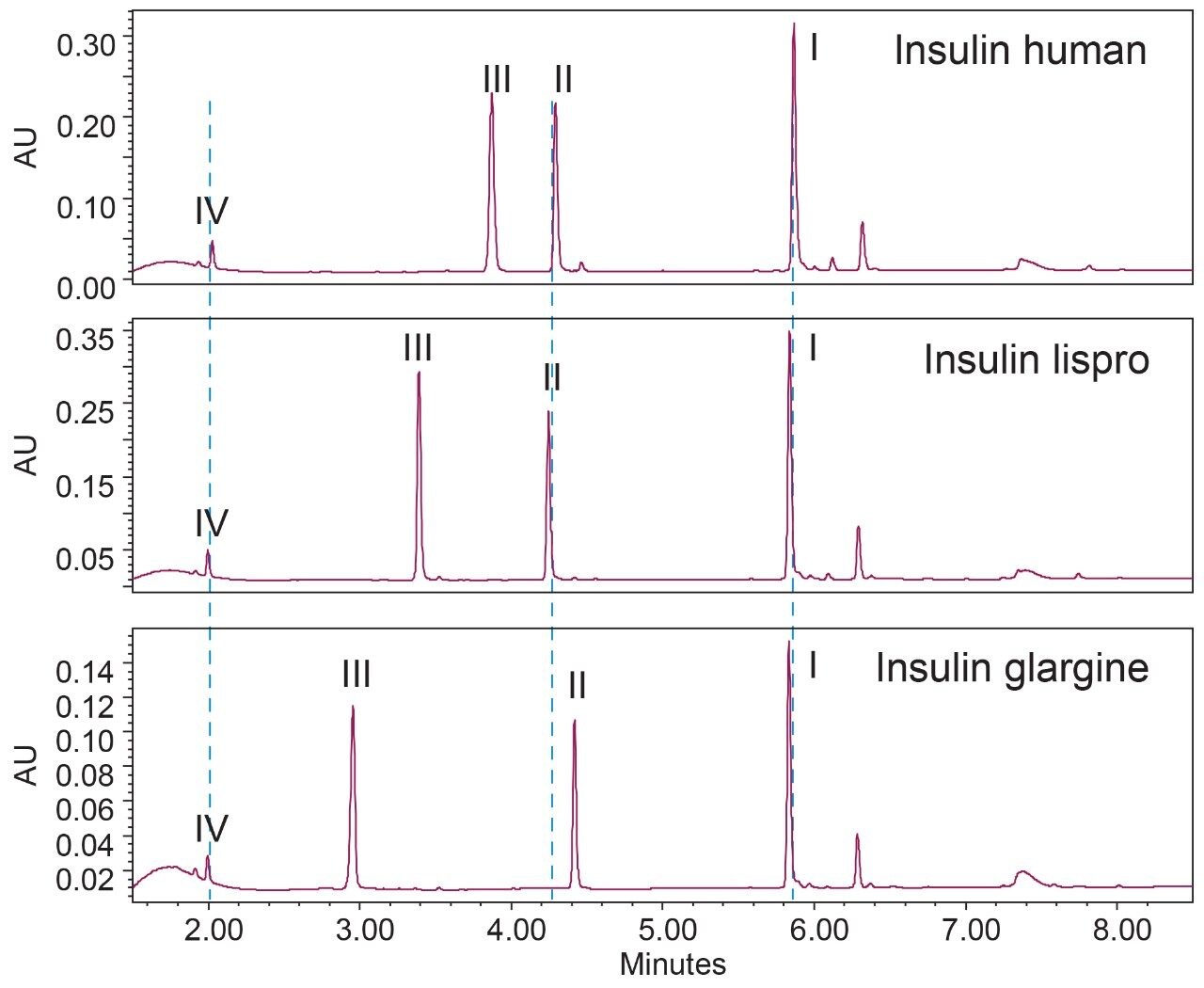
With the same method condition, the developed method was applied on insulin lispro and insulin glargine. As shown in Figure 4, the peptide fragments were well resolved for all three insulin analogues in an 8 min elution window. The top five most abundant peaks were identified as fragment I-IV, and a pyroglutamic form of fragment I. The same retention time was observed for peptide fragment IV and I across samples due to their identical structures. The retention time of peptide fragment III in insulin lispro and II, III in insulin glargine shifted from the peptide fragments peaks found in insulin human due to minor differences in their primary sequence (Figure 1), suggesting the method is robust and specific to various insulin analogues. Collectively, this study demonstrates ‘finger-print’-like analyses can be developed as efficient and robust platform methods used in routine analysis of insulin analogues.
This work demonstrated the modernization of the USP monograph for high-throughput identity test of insulin analogues. Using a sub-2-µm particle size column and ion-pairing reagent TFA, the HPLC-based USP method was updated to an UPLC-based method with higher efficiency for the separation of digested peptides of insulin analogues. Compared to original USP methods, the developed method reduced analysis time by 75% with comparable resolution and can be applied to multiple insulin analogues, making it an ideal platform method for insulin analysis in a routine environment.
720006136, November 2017