For research use only. Not for use in diagnostic procedures.
This application note describes a clinical research method utilizing Waters Oasis HLB μElution Plate technology for the extraction of 25OHD2 and 25OHD3 from human serum.
The demand for serum 25-hydroxyvitamin D (25OHD) analysis has increased dramatically in recent years. While the role of vitamin D in bone metabolism is well established, comparatively little research has been performed to elucidate its role in other diseases. However, considerable time, effort, and funds are being applied to randomized, prospective clinical trials that aim to better define the link between vitamin D status and a variety of diseases – such as cancers, multiple sclerosis, heart disease, and diabetes.
Vitamin D is available in two forms: the plant-derived vitamin D2 (ergocalciferol), and vitamin D3 (cholecalciferol), which is formed upon exposure of the skin to ultraviolet radiation. The accepted indicator of vitamin D status – total 25OHD [that is, the sum of 25OHD2 and 25OHD3] – has been a challenge to measure accurately because the antibodies used in some immunoassays do not have 100% co-specificity for both 25OHD2 and 25OHD3. Therefore, many clinical research laboratories have now adopted LC-MS/MS based methods for measuring total 25OHD while also allowing independent quantification of 25OHD2 and 25OHD3.
Described here is a clinical research method utilizing Waters Oasis HLB µElution Plate technology for the extraction of 25OHD2 and 25OHD3 from human serum. This method was automated on a Tecan Freedom Evo 100/4 Liquid Handler, allowing for full sample tracking from the primary tube to processed results. Chromatographic separation of extracted samples was performed on an ACQUITY UPLC I-Class using an ACQUITY UPLC BEH Phenyl Column followed by mass detection on a Xevo TQD (Figure 1).

|
System: |
ACQUITY UPLC I-Class |
|
Needle: |
30 μL |
|
Column: |
ACQUITY UPLC BEH Phenyl, 130Å, 1.7 μm, 2.1 mm x 50 mm (P/N 186002884) |
|
Pre-column filter: |
ACQUITY UPLC Column In-line filter kit (P/N 205000343) |
|
Mobile phase A: |
Aqueous 2 mM ammonium acetate + 0.1% formic acid |
|
Mobile phase B: |
Methanol with 2 mM ammonium acetate + 0.1% formic acid |
|
Needle wash solvent: |
80% Methanol(aq) + 0.1% formic acid |
|
Purge solvent: |
65% Methanol(aq) + 0.1% formic acid |
|
Column temp.: |
35 °C |
|
Injection volume: |
20 μL |
|
Flow rate: |
0.45 mL/min. |
|
Gradient: |
See Table 1 |
|
Run time: |
4.2 minutes |
|
System: |
Xevo TQD |
|
Resolution: |
MS1 (0.7 FWHM), MS2 (0.85 FWHM) |
|
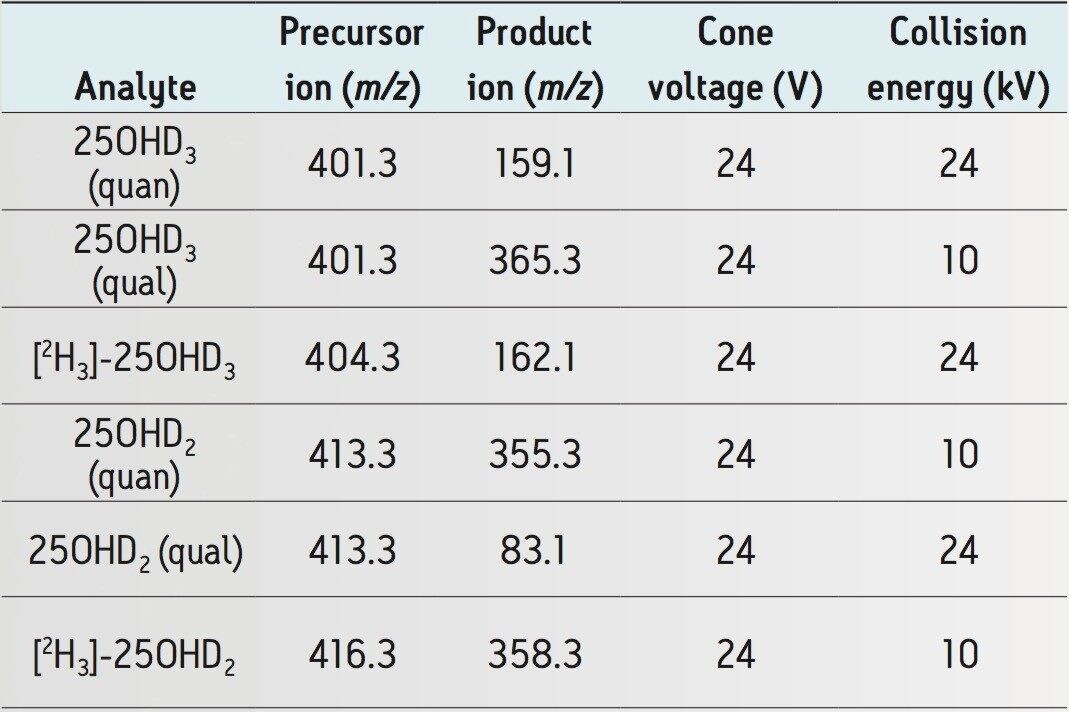
Acquisition mode: |
Multiple Reaction Monitoring (MRM) (see Table 2 for details) |
|
Polarity: |
ESI+ |
|
Capillary: |
0.80 kV |
|
Source temp.: |
120 °C |
|
Desolvation temp.: |
400 °C |
|
Dwell time: |
0.06 seconds |
|
Inter-scan delay: |
0.01 seconds |
|
Inter-channel delay: |
0.02 seconds |
MassLynx v4.1 with TargetLynx Application Manager
MassLynx Tecan File Converter v2.0
MassLynx LIMS Interface v3.0
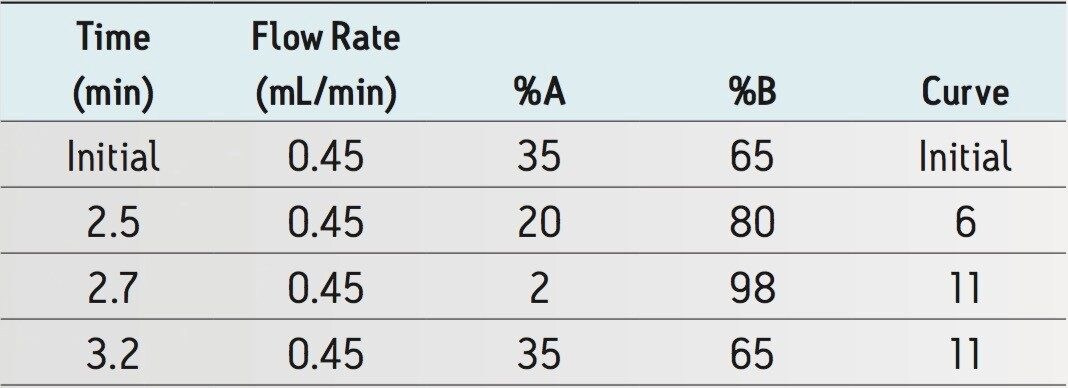
Operating backpressure at the initial conditions is ~7000 psi.
25OHD2 and 25OHD3 certified reference solutions and their stable labeled internal standards (2H3) were purchased from IsoSciences. Calibrators were prepared in a surrogate matrix of MSG2000 stripped human serum purchased from Golden West Biologicals. The calibration range for 25OHD2 and 25OHD3 was 10–375 nmol/L (~4-150 ng/mL). QC materials were purchased from Recipe at ~50 and 100 nmol/L (~20 and 40 ng/mL) and UTAK at ~25, 75, and 190 nmol/L (~10, 30, and 75 ng/mL). Distilled water, methanol, ammonium acetate, zinc sulphate, and 2-propanol were purchased from Sigma-Aldrich. Formic acid was purchased from VWR.
To convert SI units (nmol/L) to conventional mass units (ng/mL) divide by 2.423 for 25OHD2 and by 2.496 for 25OHD3.
Extraction was performed using a Tecan Freedom Evo 100/4 Liquid Handling System (LHS). Serum calibrators, QCs, and samples were placed onto the LHS along with all reagents and consumables required. Samples were identified by barcodes and tracked through the extraction procedure. 150 µL of sample was transferred into a 96-deep well plate and 20 µL of 620 nmol/L (250 ng/mL) internal standard, 150 µL 0.2 M zinc sulphate(aq), and 600 µL of methanol were added. This was followed by centrifugation (off-line) for five minutes at 750 g. The samples were mixed after each reagent addition.
The Oasis HLB µElution Plate was conditioned and equilibrated with 200 µL methanol and 60% methanol(aq) respectively. An aliquot of each pretreated sample (600 µL) was loaded into individual wells of the plate and slowly pulled through at low vacuum. Consecutive washes with 200 µL of 5% methanol(aq) and 200 µL of 60% methanol(aq) were performed. 25OHD2 and 25OHD3 were eluted using 80 µL of 95:5 (v:v) methanol–IPA, followed by 50 µL of water to match the organic strength of the initial chromatographic conditions. The collection plate was sealed manually and transferred onto the ACQUITY UPLC Sample Manager (FTN) for injection onto the UPLC-MS/MS.
A MassLynx sample list was created with the MassLynx Tecan File Converter using the barcode information scanned and tracked sample positions in the 96-well plate, which allows for complete sample tracking. The MassLynx LIMS Interface may also be used to upload sample results into a LIMS.
The sample preparation time for a full plate of 96 samples is approximately two hours, requiring minimal manual intervention:
|
T = 0 min |
Laboratory analyst locates and manually loads samples, calibrators, QCs, reagents, and consumables onto the LHS |
|
T = 30 min |
Laboratory analyst initiates the LHS pretreatment procedure |
|
T = 1 hr 15 min |
Laboratory analyst removes the protein precipitation plate for centrifugation off-line |
|
T = 1 hr 20 min |
Laboratory analyst returns the plate to the LHS and resumes the automated SPE protocol |
|
T = 2 hr 15 min |
Laboratory analyst seals the collection plate and transfers to the ACQUITY autosampler |
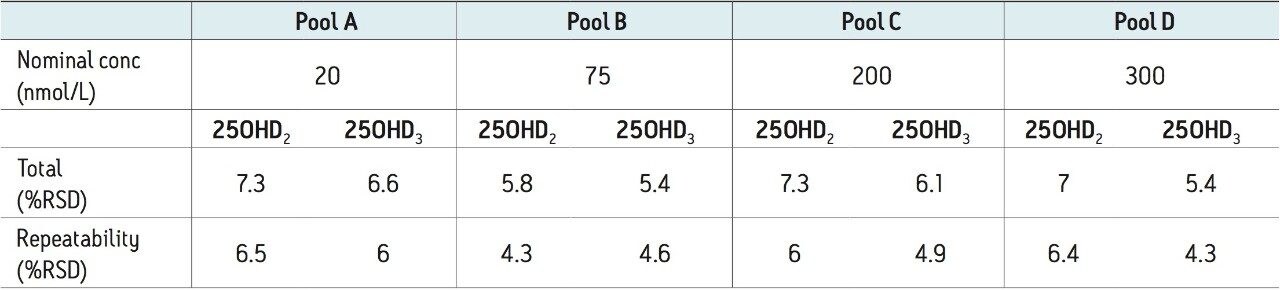
Precision was assessed by quantifying pooled human serum samples at four different concentrations in duplicate. Samples were analyzed in a fully randomized order on twenty non-consecutive days, with two analytical runs performed each day (n = 80 at each concentration level). Total precision and repeatability were ≤7.3%RSD for all runs for both 25OHD2 and 25OHD3 at concentrations between 20–300 nmol/L (8–120 ng/mL) (Table 3).
Analytical sensitivity was assessed by analyzing pooled samples that were adjusted using vitamin D depleted pooled serum to span the concentration ranges 0–15 nmol/L (0–6 ng/mL). Analysis was performed over three days with two runs per day. Using a total allowable error of 35%, the LoD and LoQ were determined to be 2.56 and 3.60 nmol/L (1.06 and 1.49 ng/mL) respectively for 25OHD2 and 4.07 and 5.56 nmol/L (1.63 and 2.23 ng/mL) respectively for 25OHD3.
The method was shown to be linear over the range of 7–450 nmol/L (2.8–180 ng/mL), when low and high pools were mixed in known ratios to give 11 samples over the range. All calibration lines in spiked vitamin D depleted serum were linear with a coefficient of determination (r2) > 0.996 over 40 occasions.
No significant carryover was observed from high concentration samples in subsequent blank samples. Recovery was assessed by supplementing each of six serum samples across the measuring range of 0–225 nmol/L (0–90 ng/mL) in addition to their endogenous levels. Samples were extracted in triplicate and the recovery for 25OHD2 and 25OHD3 ranged between 95.7%–104.6%.
An assessment of interferences was conducted by spiking the test compound at a high concentration into serum pools. Of 48 compounds tested – including endogenous and exogenous interferences, and other vitamin D metabolites – no significant interference was observed when comparing control and test sample results. Due to co-elution of C3-epi-25OHD2 with 25OHD2 and C3-epi-25OHD3 with 25OHD3, these were found to contribute to the overall 25OHD2 and 25OHD3 concentrations.
Chromatography conditions employed minimize interferences from oleamide, a known interference in sample tubes and phospholipids from the serum matrix. Below are example chromatograms of 25OHD2 and 25OHD3 samples (Figure 2).
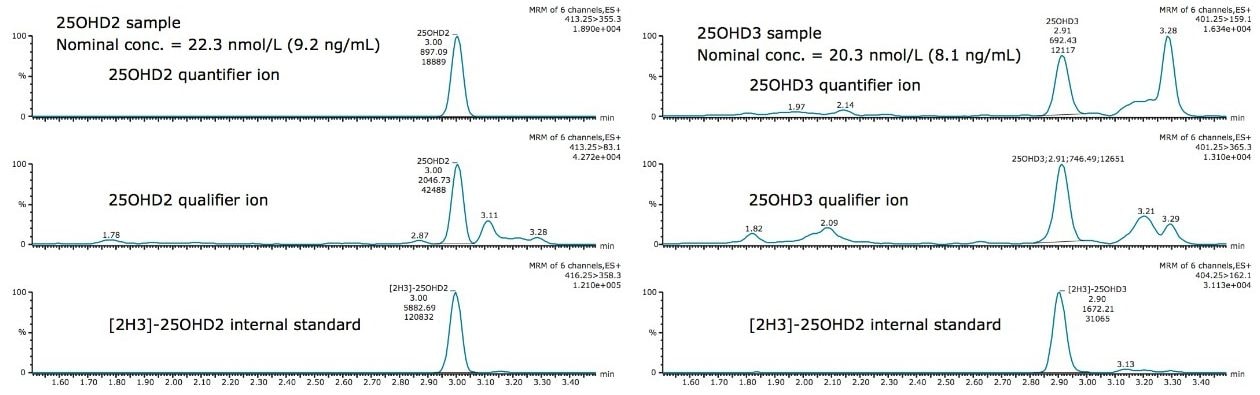
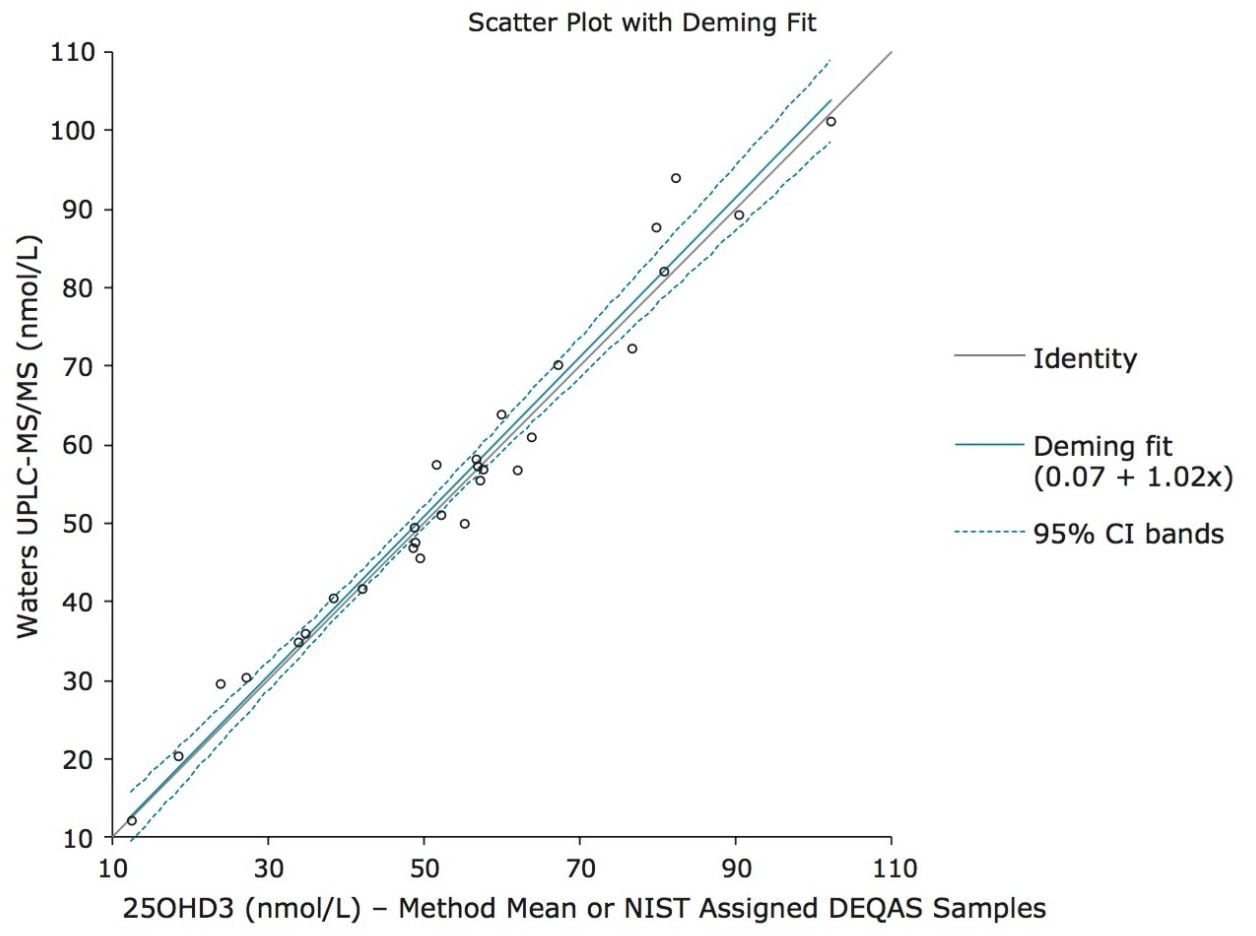
Accuracy was assessed by analyzing 29 DEQAS samples with calculated concentrations compared to the nominal value. Only six samples had a percentage difference outside of ±10%, with only one sample being outside of ±15% for 25OHD3. The comparison between DEQAS and the Waters UPLC-MS/MS method yielded a Deming regression of y = 1.02x + 0.07 (Figure 3), which showed no significant proportional or constant bias (p values of 0.7164 and 0.9744 respectively).
A semi-automated clinical research method has been developed for the analysis of 25OHD2 and 25OHD3 in human serum.
The assay described demonstrates excellent precision over 20 non-consecutive days with good linearity across the required range. A sufficient level of analytical sensitivity was achieved and no significant carryover or interferences were seen. In addition, the method demonstrates good agreement with 29 DEQAS samples.
The LHS significantly reduces the manual operation steps and operator variability to ensure more consistent and reproducible results. Furthermore, the use of Oasis µElution Plate technology eliminates the need for time-consuming solvent evaporation and reconstitution steps, allowing for the analysis of at least 192 samples per work shift.
Complete sample tracking from the sample barcode through to reporting of the result in TargetLynx and transferring the data to a LIMS has been made possible with the use of the MassLynx Tecan File Converter and MassLynx LIMS Interface.
The described semi-automated method overcomes many limitations of current LC-MS/MS method. In particular, several time-consuming and labor-intensive manual sample pretreatment steps have been eliminated. This will enable a wider range of laboratories to implement UPLC-MS/MS methodology for 25OHD analysis in clinical reasearch.
720005614, March 2016