For research use only. Not for use in diagnostic procedures.
This is an Application Brief and does not contain a detailed Experimental section.
This application brief demonstrates to attain precise large scale label-free analysis of high-density proteomic LC-MS data combined with a qualitative protein strategy that can confidently identify proteins and software to accurately quantify proteins with high reproducibility.
The depth that a complex sample can be interrogated with minimal technical variation is crucial as this characterizes the lowest abundance limit of proteins that can be both identified and quantified. This is of equal importance for the software that analyzes LC-MS data since it ultimately defines protein and peptide false discovery rates. An integrated workflow involving ProteinLynx Global SERVER (PLGS) and Scaffold Software (Proteome Software, Portland, OR, U.S.) provides protein identifications with increased confidence at low false discovery rates.
Informatics for visualizing and validating complex peptide data was successfully utilized for the qualitative analysis of Escherichia coli.
Large scale LC-MS-based discovery experiments investigating increasingly complex proteomic samples are conducted to assess variation, either experimental or biological, profile samples or quantitatively gauge differences in protein abundance. This not only places a demand on the performance of the analytical LC-MS system but also on the bioinformatics software required to analyze the data. Means of visualizing and validating data, using parameters such as probability and confidence scores, allows biological relevance to be assessed for large scale studies. Results from the SYNAPT G2-S for differentially spiked protein standards in a complex cell lysate of Escherichia coli are used to demonstrate the application of the computational and statistical tools of Scaffold Software for data independent HDMSE experiments.
A 100-ng Escherichia coli sample was injected on-column. The peptides were separated and analyzed using a nanoACQUITY UPLC System coupled with a SYNAPT G2-S operating at a precursor and product ion mass resolution of >20,000 FWHM, and data acquired in LC-HDMSE scanning mode. Searches were conducted with ProteinLynx Global SERVER v. 3.0 and visualized using Scaffold v. 3.6.
Figure 1 shows the plug-in export tool of ProteinLynx Global SERVER capable of automatically creating one or more Scaffold compatible data files. Following the upload of the search results, protein identifications can be reviewed within Scaffold using the Samples view. Criteria such as minimum protein probability, minimum peptide number, and minimum peptide probability can be useful criteria for filtering identifications. Figure 2 represents Escherichia coli identifications that were filtered accordingly, along with associated gene ontology (GO) terms to define biological function of the protein(s). Sequence information can be obtained by reviewing fragmentation data.
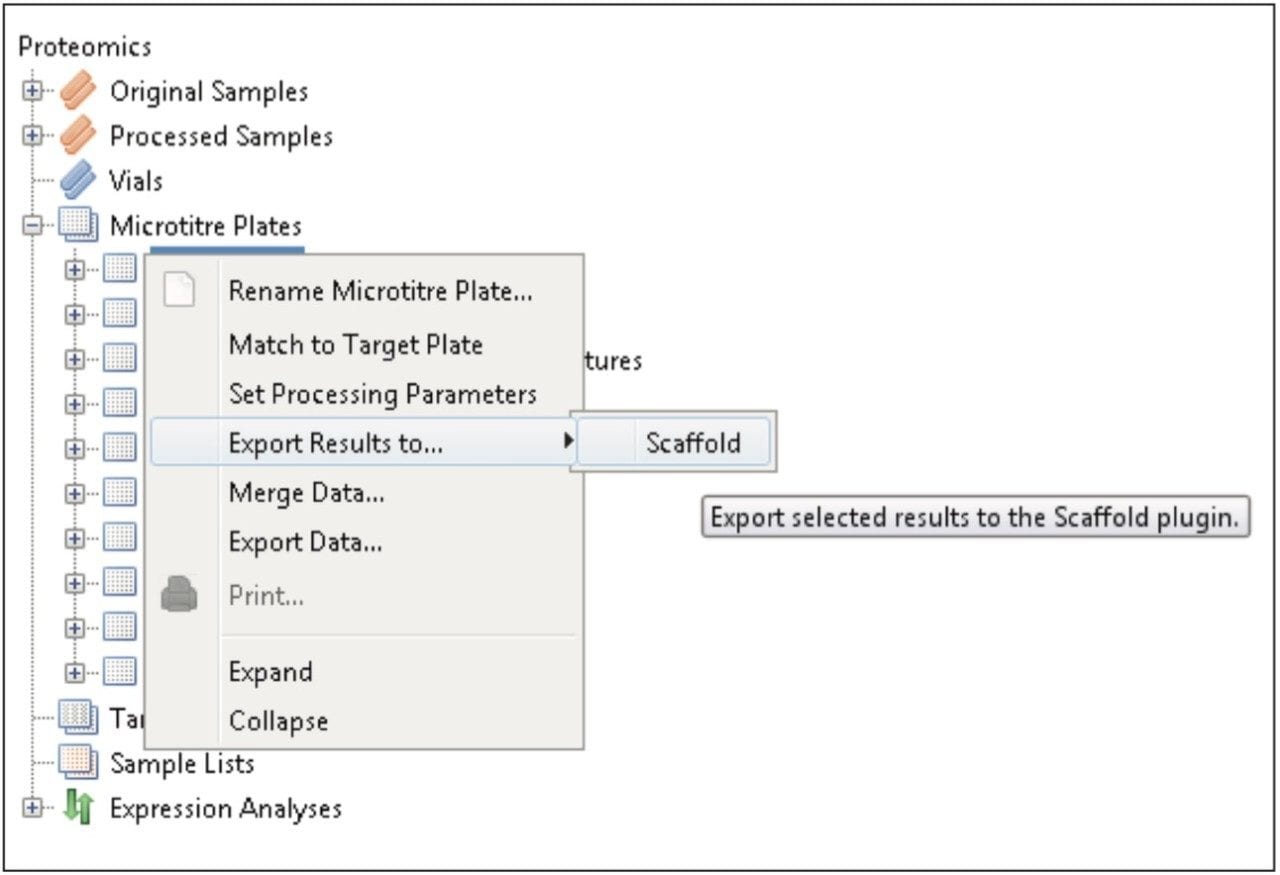
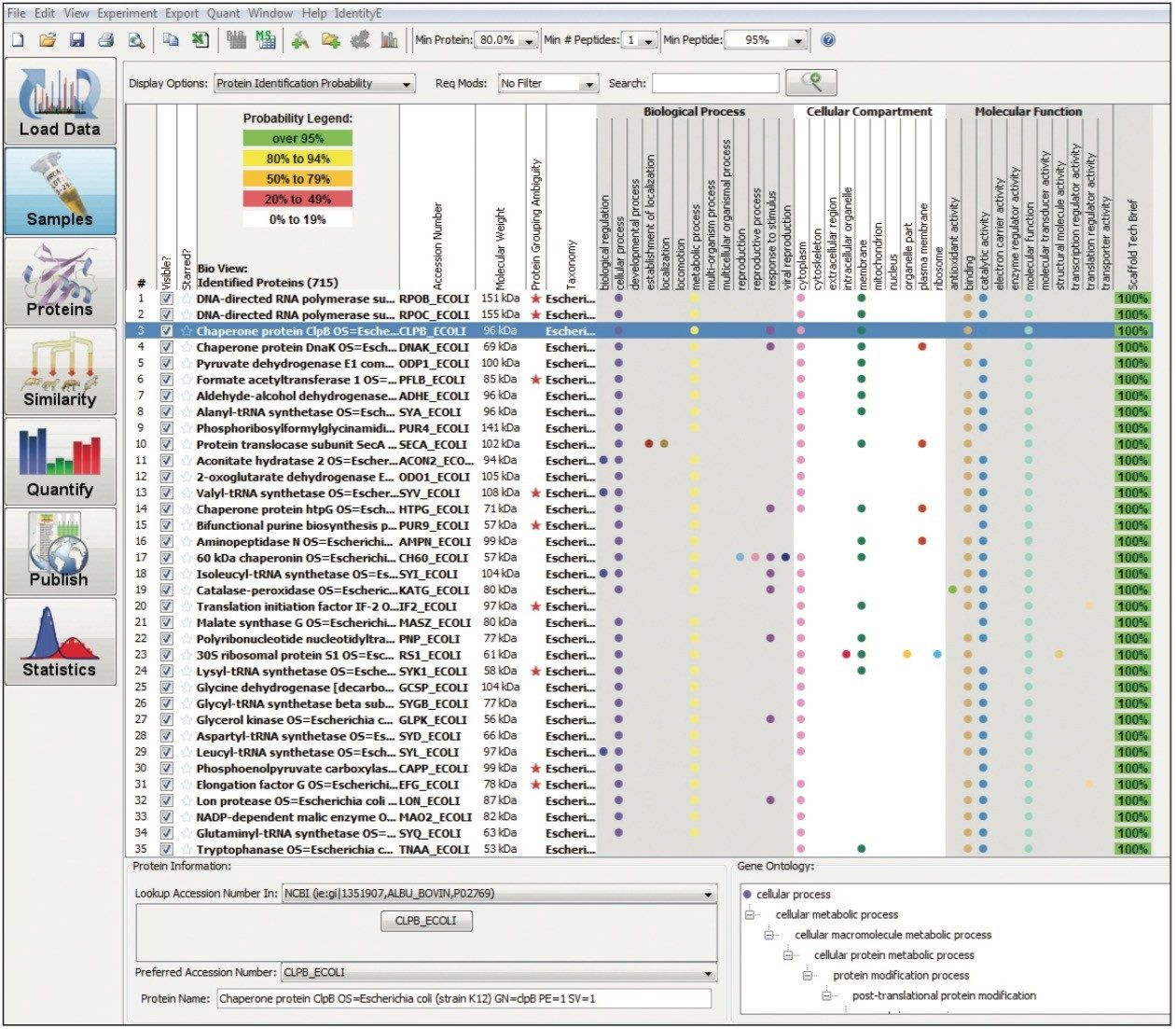
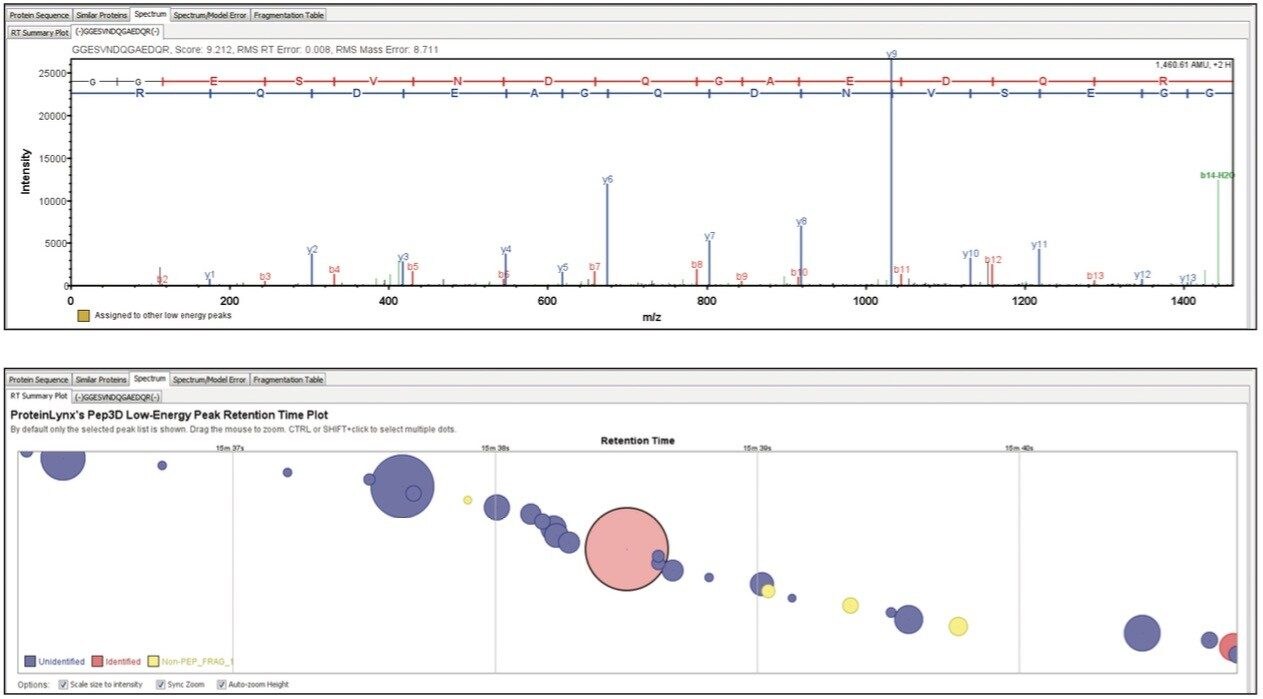
Figure 3 shows the spectrum of a peptide originating from the CLPB chaperone protein. Full sequence information is provided, thereby providing a high degree of confidence to the identification. Additional retention time information relating to the peptide of interest can be visualized from the low energy data, illustrating other species detected close to the chromatographic apex or (partially) co-eluting with the peptide of interest.
Qualitative analysis of Escherichia coli has been demonstrated using a SYNAPT G2-S operating in LC-HDMSE mode of acquisition (LC-IM-DIA-MS). Data were processed using ProteinLynx Global SERVER with the results reviewed and visualized using Scaffold Software.
720004599, February 2013