
This is an Application Brief and does not contain a detailed Experimental section.
This application brief demonstrates to accurately and efficiently identify and quantify pesticide residues present at MRL concentrations in a mandarin sample extract. Here we show results generated using the new UNIFI Scientific Information System.
The Pesticide Screening Application Solution has been custom built to confidently report and quantify pesticide residues using a streamlined workflow.
Current trends regarding pesticide use indicate that more than 500 compounds are routinely used on a global basis with different countries having varying regulations concerning licensing and legislative limits. With increasing global trade there is a requirement for multi-residue screening strategies capable of efficiently detecting residue violations to ensure consumer safety. Using the Waters Pesticide Screening Application Solution, analysts can meet these challenges in both food and environmental matrices. UNIFI is a revolutionary new scientific information system that enables scientists to test and report more samples for an increasing number of residues and contaminants. UNIFI captures complex UPLC-MSE data in a single platform and then rapidly processes the data to confirm and quantify the presence of pesticide residues. The Xevo G2-S QTof has high mass accuracy, exceeding the performance criteria specified in SANCO guidelines1 of 5 ppm. Using a scientific library, that includes molecular formulae, fragment ion and retention time information, we were able to confidently detect low concentrations of pesticide residues following a simple extraction protocol.
Mandarin extracts were kindly supplied by WIV-ISP for this study. The chromatographic separation was performed using an ACQUITY UPLC I-Class System with a water/methanol gradient. The data were collected using the Xevo G2-S QTof in MSE mode, where both precursor ions and fragment ions are acquired in a single injection. Matrix-matched standards were prepared with 116 pesticides to generate a calibration series equivalent to 0.001 to 0.1 mg kg-1 in mandarin. Over 100 pesticides were spiked into a blank mandarin extract and a non-targeted analysis was performed against a subset of 479 pesticides from the scientific library.
Processing simultaneously acquired accurate mass precursor and fragment ion (MSE) data using UNIFI enabled the confirmation of pesticide residues with low false positive rates (≤5%). The enhanced quantitative performance of Tof technology, shown in Figure 1, is clearly demonstrated by the calibration series for imazalil. A correlation coefficient (R2) of 0.999 was obtained along with a limit of detection of 0.001 mg kg-1 in mandarin. Additional functionality provided by UNIFI, including a total component summary, component details, and component plots are also shown. The component information can be easily accessed and displayed in a variety of user-defined views, providing the scientist with added confidence in the results.
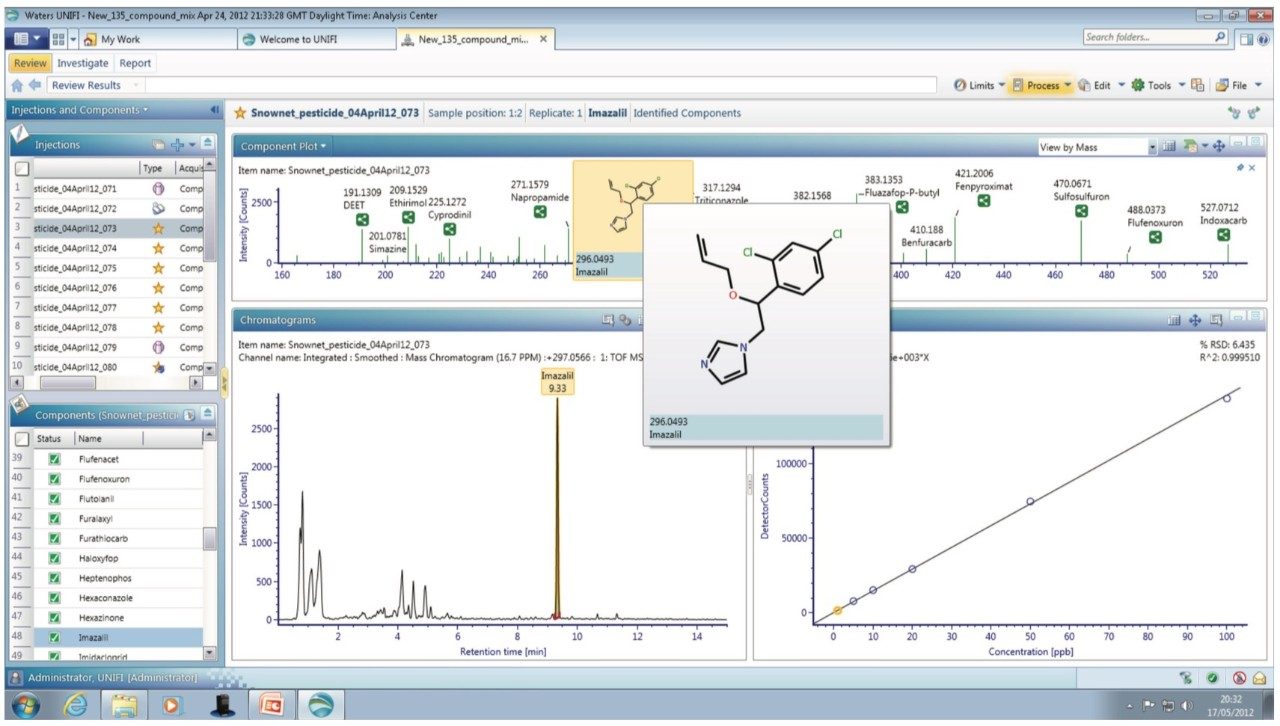
Analyte specific information that can be used for confirmation purposes, such as molecular structure, adduct formation, and fragmentation, can be generated from the accurate mass (≤2 ppm) data. The MSE spectra generated for metalaxyl is shown in Figure 2. The component summary view shows all the components identified in the mandarin sample extract. The information-rich MSE data, in combination with the UNIFI scientific library allow the pesticides to be identified based on accurate mass of both precursor and fragment ions, isotopic pattern, and retention time information. This enables pesticide residues to be confirmed with a high degree of confidence and minimizes false positives.
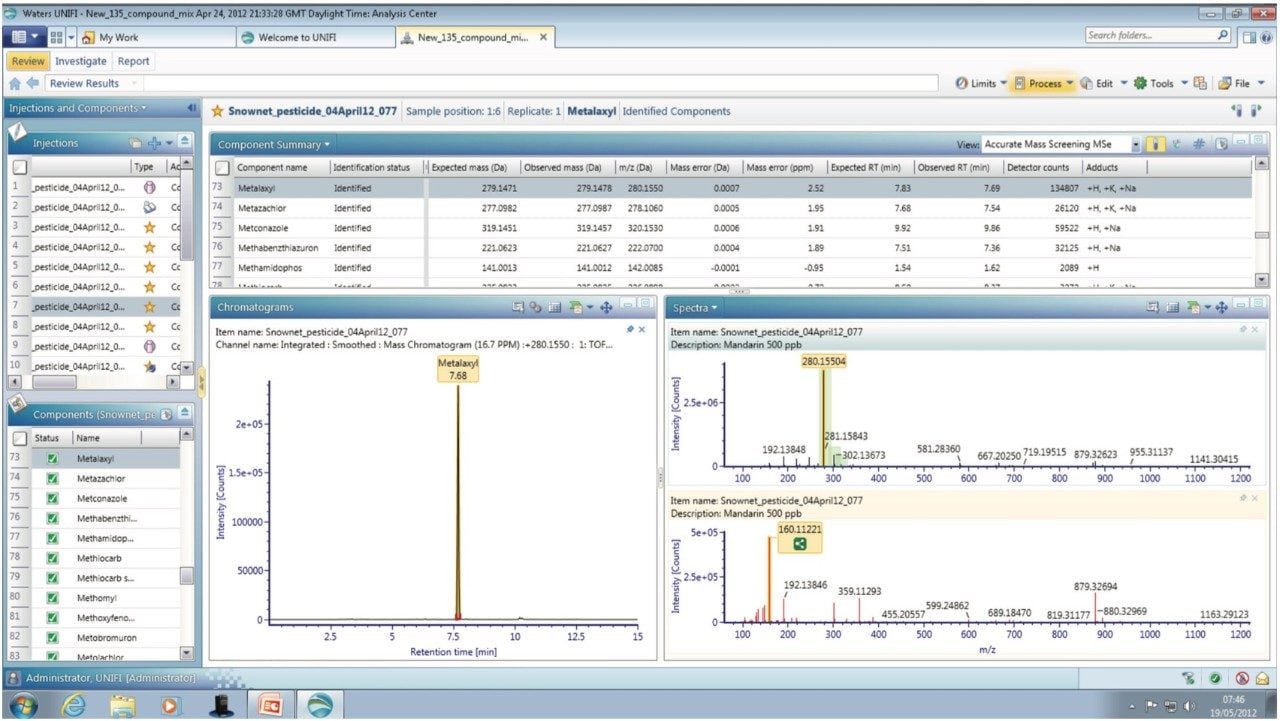
In this technology brief we have demonstrated the functionality of the new Pesticide Screening Application Solution for routine detection, quantification, and reporting of pesticide residues at the relevant regulatory limits in complex food matrices. The UNIFI Scientific Information System uses the accurate mass of both precursor and fragment ions, isotopic pattern and intensity, and retention time information to reliably detect and confirm pesticide residues while minimizing false positive and negative results (≤5%), in accordance with EU guidelines.
720004406, June 2012