
In this application note, we demonstrate the use of the Waters SYNAPT G2 HMDS System to provide high resolution and enhanced mass measurement accuracy across a wide range of concentrations, at scan rates compatible with UPLC separations. This system provides the highest levels of specificity for rapid structural elucidation of drug metabolites from complex in vivo samples.
We highlight the impact of combining enhanced mass accuracy and precision with MassFragment Software, which automates fragment ion spectral interpretation. Scientists can now move through the structural elucidation process more rapidly and confidently than previously possible.
Metabolite identification is an integral part of discovery and development programs within pharmaceutical companies of any size. One of its most significant bottlenecks is the structural elucidation process. Traditionally this process has been conducted with nominal mass instrumentation such as tandem quadrupole and ion trap mass spectrometers, with time-consuming manual interpretation of the spectral data. In recent years, there has been a shift to the use of platforms that are capable of high resolution and exact mass measurement (e.g., time-of-flight (Tof) or quadrupole time-of-flight (QTof) mass spectrometry), since their additional levels of specificity reduce falsepositive identifications. Such technologies also improve the process of characterizing complex fragmentation pathways.
The SYNAPT G2 HDMS System delivers a paradigm shift for the metabolite identification process due its ability to combine the full power of UPLC separations,1 high-resolution mass spectrometry, and intelligent informatics to significantly reduce or eliminate false positive results in the structural elucidation process.
SYNAPT G2 HDMS, with its breakthrough quantitative Tof technology, QuanTof, provides up to 40,000 FWHM, sub-1-ppm RMS mass accuracy, enhanced mass precision, and enhanced dynamic range (>104) – all at acquisition rates of up to 20 spectra/sec. This is unlike FT-MS or electrostatic ion trap-based MS systems whose resolution and dynamic range reduces significantly as spectral acquisition rate and sample complexity respectively increase.
SYNAPT G2 HDMS collects comprehensive fragment ion (MS/MS) information from every detectable molecular ion across the entire chromatographic separation provided by UPLC.
MassFragment,2 an intelligent software tool, automates structural assignment to fragment ion spectra, making data processing significantly easier.
|
Sample: |
Rat plasma dosed with ritonavir at a concentration of 10 mg/kg |
|
LC system: |
Waters ACQUITY UPLC System |
|
Column: |
ACQUITY UPLC HSS T3 2.1 x 100 mm, 1.8 μm |
|
Flow Rate: |
0.5 mL/min |
|
Injection vol.: |
40 μL |
|
Mobile phase A: |
5 mM Ammonium Acetate pH 5 |
|
Mobile phase B: |
Acetonitrile |
|
MS system: |
Waters SYNAPT G2 HDMS System |
|
Acquisition mode: |
UPLC-MSE |
|
Ion mode: |
ESI positive ion mode |
|
Cone voltage: |
30 V |
|
Capillary voltage: |
2.8 kV |
|
Desolvation temp.: |
450 °C |
|
Source temp.: |
120 °C |
|
Collision energy: |
Trap 20 eV, Transfer 20 eV |
|
Collision gas: |
Argon |
|
Lock mass: |
Leucine enkephalin, m/z 566.2771, 1 ng/μL at a flow rate of 50 μL/min |
|
Data management: |
MassLynx Software with MassFragment Software |
MassFragment Software utilizes systematic bond disconnections and a scoring system that depends on the type of bond disconnected. For instance, breaking a phenyl bond – which is not very likely – would result in the assignment of a high score of 8. In general, the lower the score and the more accurate the MS data, the more likely the assignment is correct. These scores are also customizable, providing scientists with flexibility to manage the result evaluation process. The overall result is a structural representation of the fragment ion together with its exact mass, elemental composition, and doublebond equivalence.
The key to streamlining the results evaluation process is data with high mass accuracy and precision obtained with the SYNAPT G2 HDMS System. Figure 1 demonstrates this capability with a narrow mass error window of 0.5 mDa, which can be used to assign the most feasible structure(s) to each fragment ion within an MSE or MS/MS spectrum. Very narrow mass window of +/- 0.5 mDa Minimizes false positives and speeds up structural identification Figure 1.
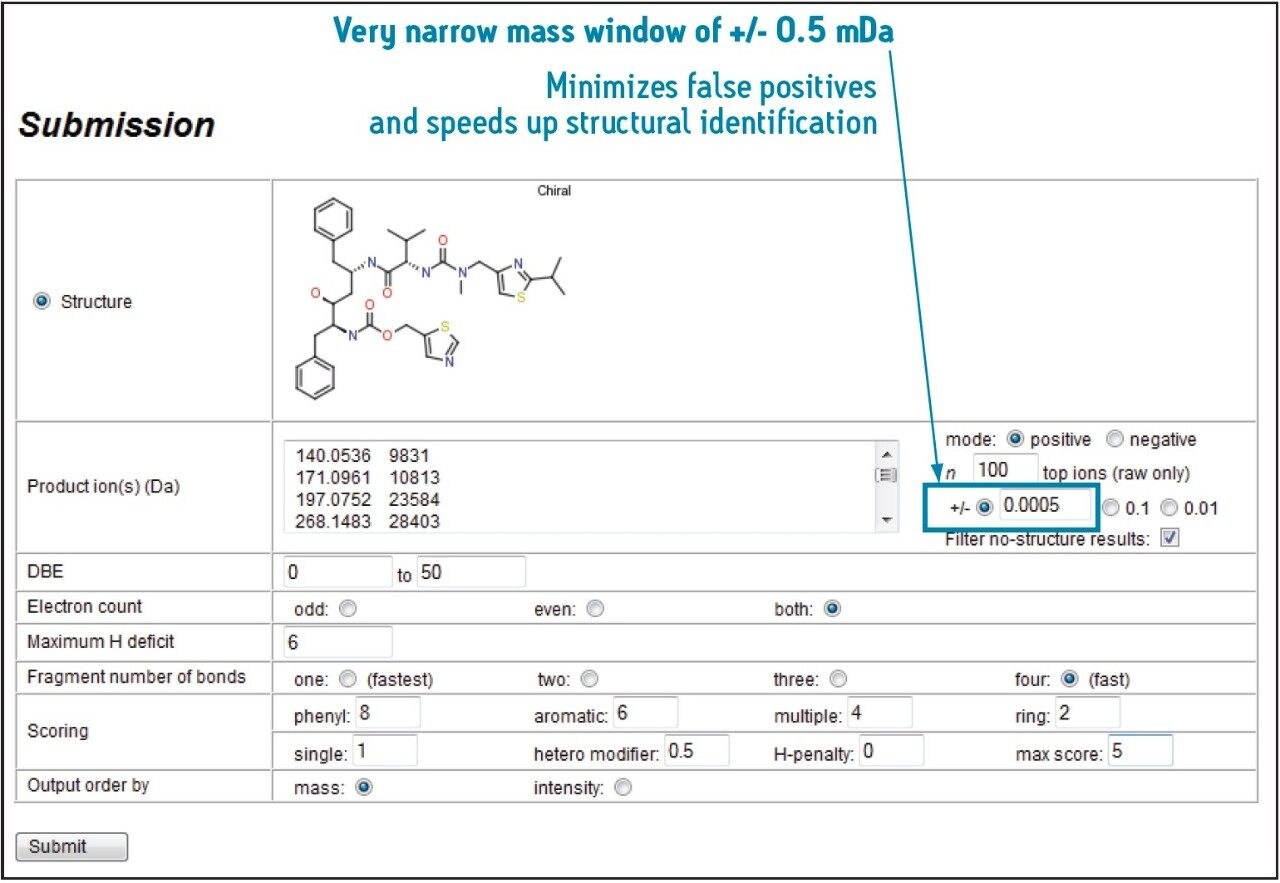
MassFragment automatically extracts fragment ion spectra from MassLynx Software or its specialized MetaboLynx XS Application Manager for submission to the structural assignment process, as displayed in Figure 1.
The outstanding mass accuracy and precision provided by the SYNAPT G2 HDMS System – defined as the minimized variation in mass error from scan to scan across the chromatographic peak – provides the highest levels confidence in results, as shown in Figure 2. In this particular analysis, MassFragment proposed only one possible structure for each of the fragment ions of the parent drug ritonavir, thus leading to unambiguous structural elucidation of the drug.

Once all the fragment ions for the parent drug have been confidently identified, it is logical to then use this information to interpret the fragment ions of other metabolites. The ability to quickly and confidently identify common fragment ions with the parent drug is very important since it will enable more effective localization of biotransformations (i.e., the portions of the molecule that have variable structure), which can be confirmed using exact mass measurements. This is clearly observed in Figures 3 and 4, where the mass accuracies for both the parent drug and the hydroxylated metabolite of ritonavir are all within 0.5 mDa for all fragment ions.
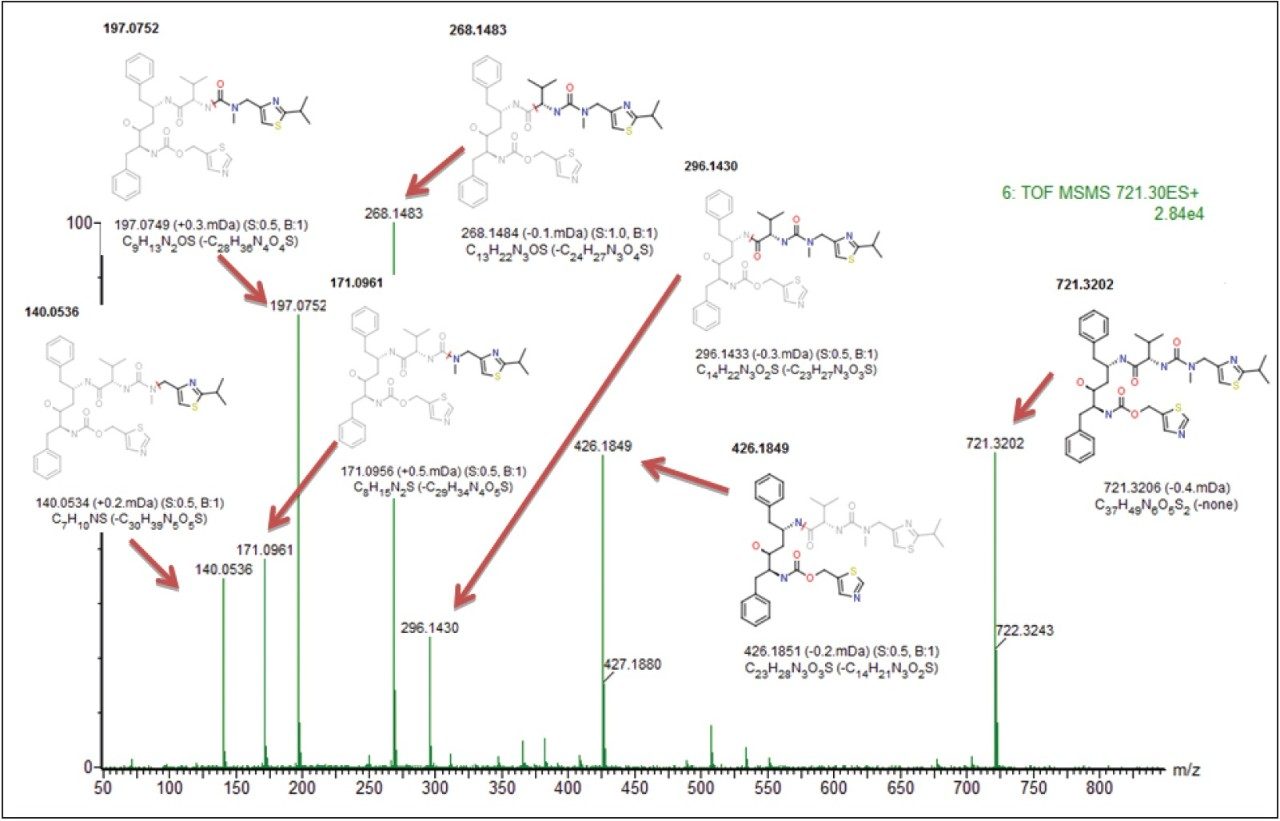

The combination of the SYNAPT G2 HDMS System’s high mass accuracy and precision with intelligent MassFragment Software provides a fast, efficient, and unambiguous route to assigning structure to fragment ion spectra.
This comprehensive and intelligent analytical workflow can have a dramatic impact on productivity levels where LC-MS/MS is used for routine structural elucidation in metabolite, lipid, or impurity profiling applications.
720003183, August 2009