Application Brief
This is an Application Brief and does not contain a detailed Experimental section.
Matthew E. Daly1, Matt Spick2, Christopher J. Hughes1, Scarlet Ferrinho1, Cheryl M. Isherwood3, Lee A. Gethings1, Hana Hassanin4, Daan R. van der Veen3, Debra J. Skene3, Jonathan D. Johnston3
1 Waters Corporation, USA
2 School of Health Sciences, Faculty of Health and Medical Sciences, University of Surrey, Guildford, UK
3 Section of Chronobiology, School of Biosciences, Faculty of Health and Medical Sciences, University of Surrey, Guildford, UK
4 Clinical Research Facility, Faculty of Health and Medical Sciences, University of Surrey, Guildford, UK
Published on April 30, 2025
This is an Application Brief and does not contain a detailed Experimental section.
For research use only. Not for use in diagnostic procedures.
Clinical proteomics and large cohort studies rely on high throughput workflows, enabling high quality results to be generated quickly. However, these may be compromised by the sensitivity of the assay, in addition to other factors of the experiment, relying on highly abundant proteins being identified as potential biomarkers. Therefore, high throughput, high flow rate experiments may not be sufficient to identify low abundant proteins that may act as highly significant biomarkers. The Evosep One can help bridge this gap due to its standardized microflow chromatography, and indeed when coupled to a SYNAPT™ XS Mass Spectrometer, the system can be readily utilized for high throughput analyses. This study involved the analysis of undepleted human sera collected from 24 healthy male patients, collected at 30-minute intervals over the course of 30 hours. The total analysis consisted of over 480 injections, which resulted in eight days of instrument acquisition time. The circadian rhythm of the circulating serum proteome was characterized with high reproducibility and confidence, resulting in the identification of 281 proteins (based on triplicate replicates) allowing for downstream analysis and identification of temporal variations in the human plasma proteome.
Circadian rhythm is a natural endogenous rhythm within many organisms repeating approximately every 24 hours and affecting genetic, metabolic, hormonal, and behavioral signals. Within humans, the rhythm is controlled by the suprachiasmatic nucleus within the hypothalamus of the brain which, via two transcriptional-translational feedback loops, has a downstream effect on gene transcription.1 The circulating plasma proteome therefore, demonstrates changes in terms of both abundance and the level of post-translational modification during natural circadian rhythm that can be characterized by mass spectrometry.2 Characterization of these changes can offer insights into disorders associated with the dysregulation of circadian rhythm with the possibility of identifying potential biomarkers.
However, identification of such potential biomarkers necessitates the analysis of large study cohorts such that there is sufficient statistical power to produce markers with high predictive value. Therefore, a high-throughput scalable method whilst maintaining proteome coverage is important when designing such experiments.
The Evosep One is a standardized chromatographic platform employing pre-defined gradients tailored to the number of samples required to be analyzed per day (e.g. 60 sample per day; 60 SPD).3 The sample is eluted from a disposable trap column and the analytical gradient is pre-formed by low pressure-pumps and placed into a gradient loop, where through valve switching, the sample is introduced to and eluted from the analytical column using the pre-formed gradient and one high-pressure pump. This two-pump approach acts to increase pump lifetime whilst maintaining the sensitivity increases from nanoflow or low microflow separations. Serum samples from 24 patients were taken across a 30-hour time course resulting in a total of 1440 individual patient/timepoint snapshot samples.4 For analysis of this large cohort, the Evosep One was coupled to a SYNAPT XS Ion Mobility Mass Spectrometer (Figure 1). Due to compression of the chromatographic gradient and consequently, eluting peaks, the ion mobility-enabled SYNAPT XS allowed for increased peak capacity, effectively deconvolving co-eluting chromatographic peaks in real-time and leading to an increase in peptide and protein identifications across the entire analysis.
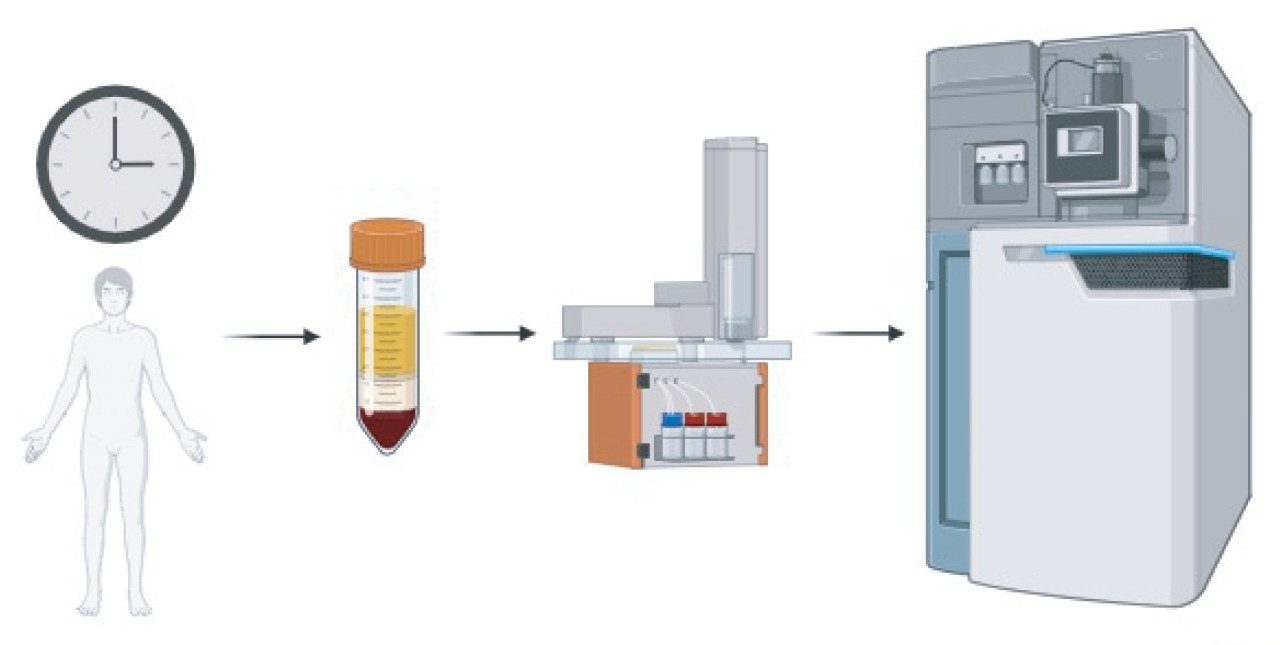
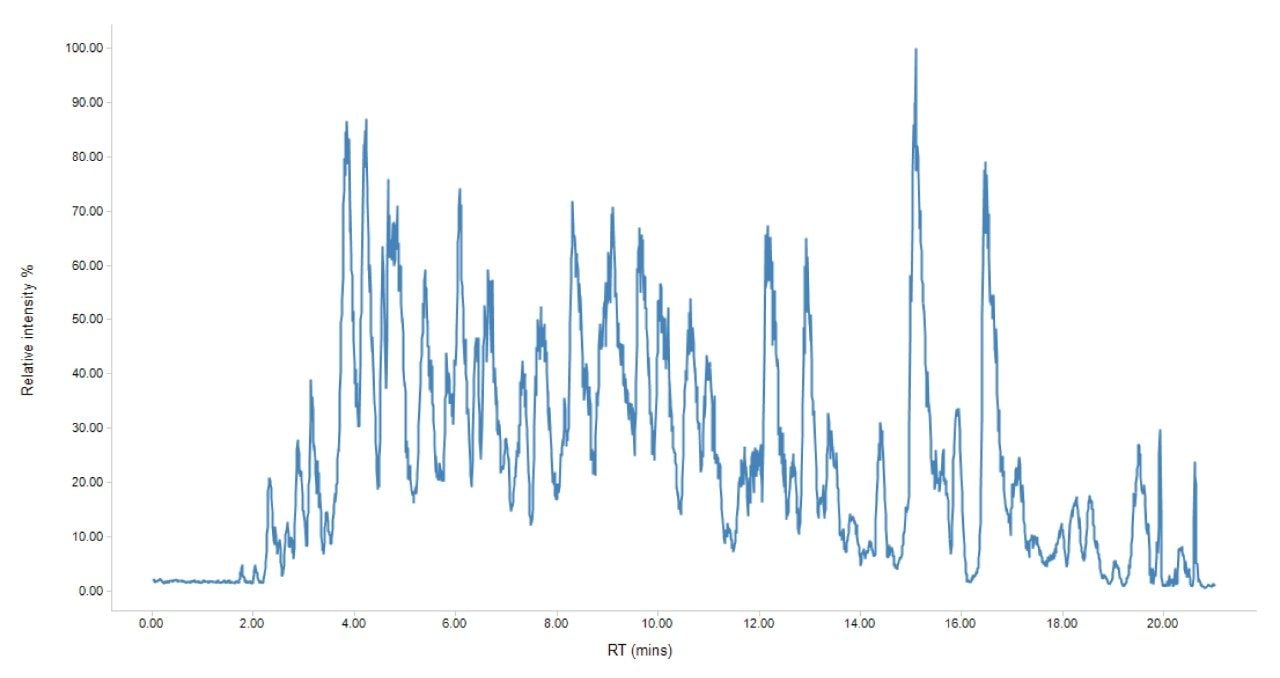
A subset of blood draws originating from 10 patients was analyzed, constituting over 160 individual samples. Each sample was reduced, alkylated and enzymatically digested with trypsin – no other sample enrichment or depletion was performed. Digested samples were prepared using the Evotips protocol which consist of a C18 resin packed inside of a pipette tip, effectively acting as a disposable trap column capable of desalting and pre-concentration of samples prior to chromatographic separation on the analytical column. Three replicates per sample were prepared using this procedure to allow for suitable statistical analysis and hence resulting in >480 prepared Evotips. Due to the large number of sample injections required, the 60 SPD protocol was utilized to provide a balance between throughput and coverage, whilst mitigating any batch effects. A representative chromatogram is shown in Figure 2.
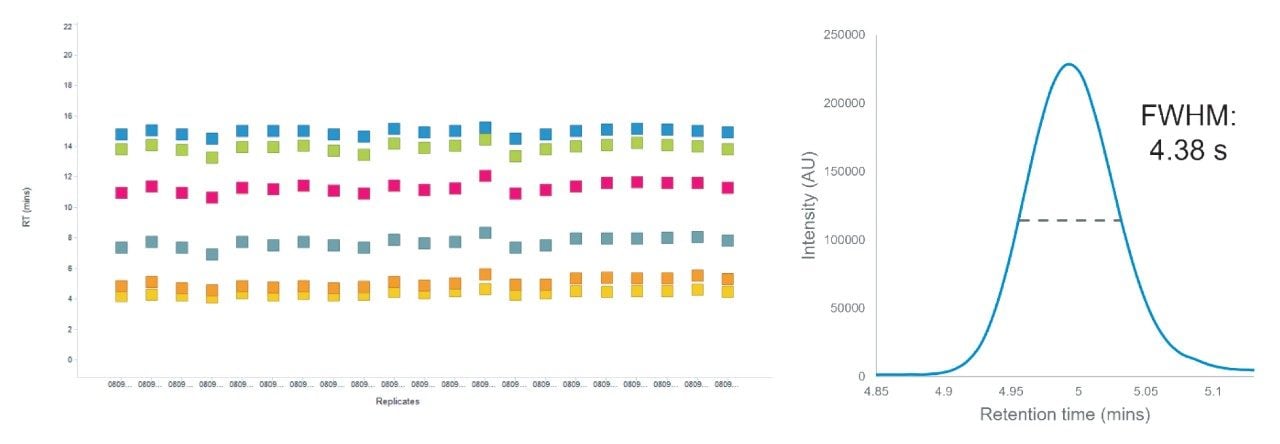
The chromatographic performance was then monitored and evaluated using a suite of marker peptides spanning the active gradient that were confidently identified in all injections (Figure 3). Of the marker peptides monitored, the measured retention time varied by less than 5% demonstrating excellent chromatographic reproducibility across >480 injections acquired over eight days. Alongside retention time, peak shape was also evaluated and demonstrated primarily Gaussian peak shape across runs and peptides. Short separation times and compression of chromatographic gradients can result in co-elution, and therefore the full width half maximal (FWHM) of eluting peptide, a metric normally employed to discuss chromatographic performance, must be as narrow as possible to facilitate downstream deconvolution. For example, the FWHM of peptide VQHIQLLQK (resulting from Fibrinogen alpha chain (UniProt #P02671) eluting at ~five minutes was shown to be 4.38 seconds, demonstrating excellent peak capacity of the column and gradient used.
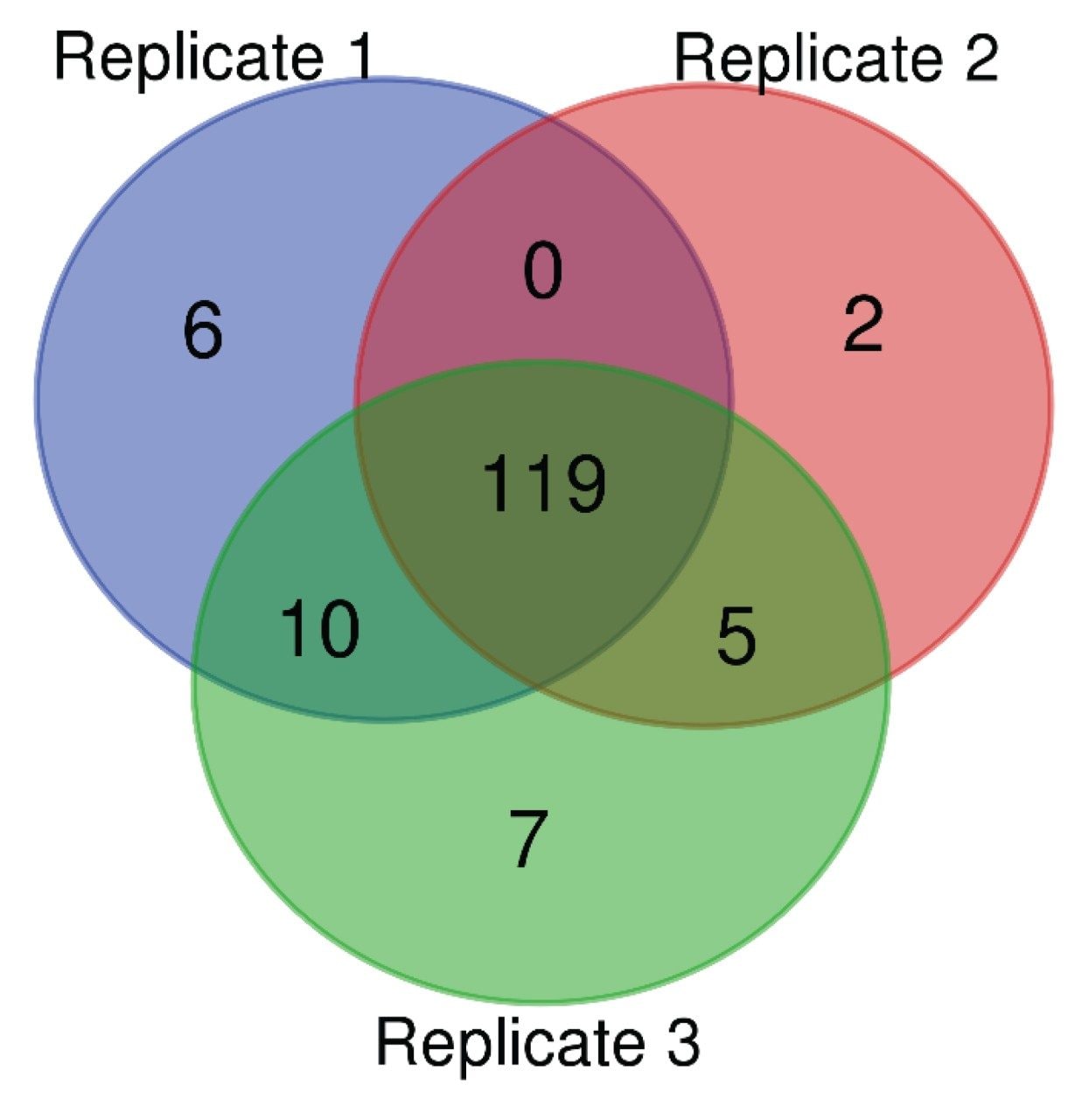
A total of 281 protein groups were identified across all sample injections aided by the ion mobility-enabled data independent mode of acquisition, HDMSE offered by the SYNAPT XS.5 The SYNAPT XS not only provided a high number of protein IDs but also demonstrated exceptional dynamic range of almost five orders of magnitude within a single sample, necessary when working with undepleted serum samples where low abundant proteins can be masked by highly abundant proteins such as albumin. Technical reproducibility was also exceptional, with over 80% of proteins identified in at least two out of three technical replicates, and 90% present in all three (Figure 4). Such technical reproducibility can be credited not only to the standardized sample preparation and chromatography, but also to the robustness and stability of the mass spectrometer.
The overall purpose of the study was to identify changes in protein expression linked to circadian rhythm, as such, the abundance of different proteins was tracked across the time course samples.4 Initially, thirteen proteins were identified as demonstrating abundance changes linked to either a 24- or 12-hour rhythm. Some example proteins that exhibited large abundance changes were the apolipoproteins (Figure 5, >25% RSD) and indeed these proteins were identified as having either a 24- or 12-hour rhythm which is understandable due to their function of affecting lipoprotein metabolism which varies throughout the day. Overall, according to analysis using Gene Ontology and Biological processes, the proteins identified as either circadian or ultradian were involved with plasminogen activation and negative regulation of triglyceride metabolism.
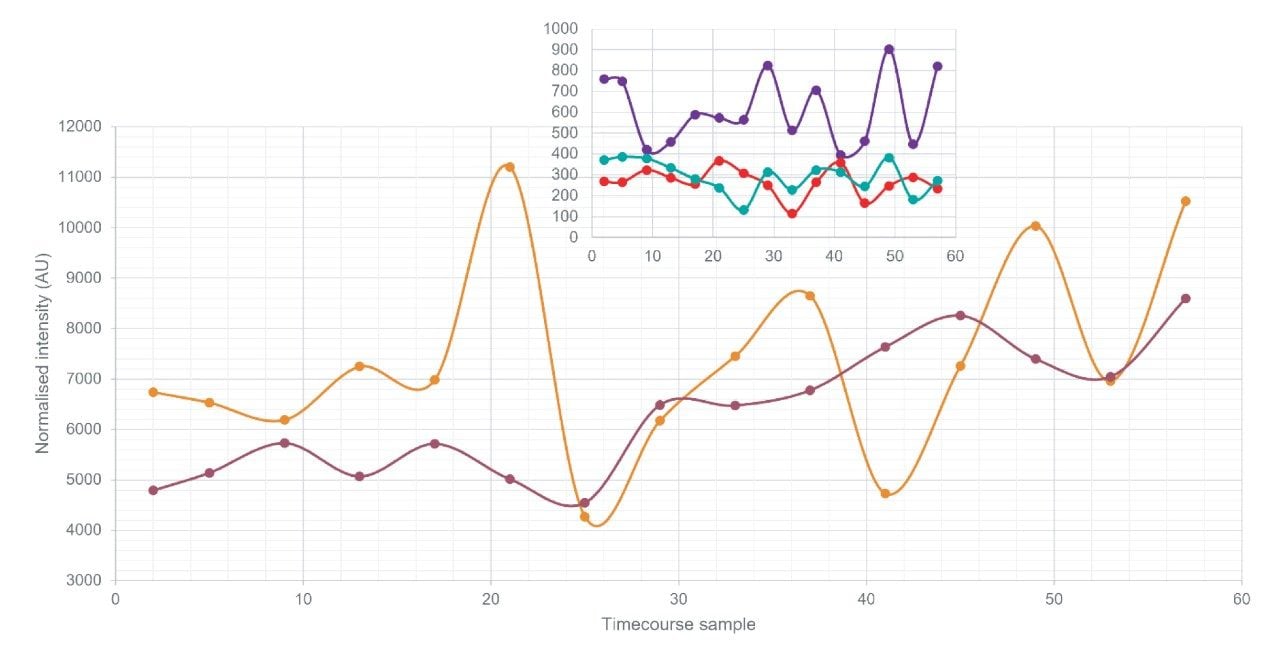
Large scale proteomic studies that contain hundreds of samples have previously compromised between longer, more sensitive nanoscale chromatography or higher-throughput standard flow methodologies. It has been demonstrated that the Evosep One utilizing standardized sample preparation and chromatographic conditions in tandem with the high peak capacity capabilities of HDMSE on the SYNAPT XS facilitated analysis of a large sample set of sera samples with high technical reproducibility. Excellent retention time stability alongside narrow peak widths observed from the Evosep One assisted in the identification of over 280 protein groups from undepleted digested serum. Several different proteins were identified with high abundance changes across the time course, in particular the apolipoproteins varied by >25%. Furthermore, although undepleted, many low abundant proteins could still be reliably identified in part due to the high dynamic range provided by the SYNAPT XS.
With ever increasing study sizes, reproducible, robust, high-throughput chromatography coupled to a mass spectrometer with sufficient peak capacity to sufficiently sample the narrow chromatographic peaks is needed to ensure high quality data is generated within acceptable time frames. The benefits of the Evosep One in combination with the SYNAPT XS is highlighted, allowing the acquisition, processing and identification of potential proteomic biomarkers with high confidence in timescales that are amenable for high throughput studies/laboratories.
720008734, April 2025