Quantification of a Lipid Conjugated Antisense Oligonucleotide (ASO) Extracted From Rat Plasma Using the OligoWorks™ SPE Microplate Kit on a HRMS System
Abstract
Discovery bioanalysis laboratories play a critical role in the drug discovery process. These laboratories are tasked with supporting a large number of potential candidates for each project, are bound by tight timelines and must be diligent and efficient in their screening process to ensure rapid decision making. As such, these laboratories rely on standardized analytical workflows to facilitate robust sample preparation and quantitative LC-MS workflows, to minimize method development times. In this work, we highlight the extraction and high recovery of a lipid conjugated ASO from rat plasma using the OligoWorks SPE Microplate Kit, along with a generic HRMS data acquisition method that maximizes productivity without compromising data quality.
Benefits
- The OligoWorks SPE Microplate Kit provides a standardized, detergent-free kit-based solution for the extraction and LC-MS quantification of therapeutic oligonucleotides from biomatrices with minimal method development required
- The Xevo™ G3 QToF HRMS instrument allows sensitive, robust and accurate low-level detection and quantification of oligonucleotide therapeutics; facilitating efficient screening in discovery and early development labs
- A simplified and streamlined workflow that can be adopted and implemented across discovery bioanalysis laboratories
Introduction
Oligonucleotide Therapeutics (ONTs) are a key focus area for many drug developers today given their powerful ability to address disease biology at the level of gene transcription and translation, and for their high target specificity and low toxicity. As the pipeline for this class of nucleic acid therapeutics continues to expand, so does the need for sensitive, accurate and robust bioanalytical assays to support their discovery and advancement through each stage of development. Discovery bioanalytical laboratories strive to streamline most aspects of analytical testing to increase efficiency as they are required to screen large numbers of potential candidates. A standardized sample preparation approach that works across a wide variety of oligonucleotides can be extremely useful in this regard and can save significant method development time and cost in these laboratories. Additionally, high resolution mass spectrometry (HRMS) is often used to collect data in an untargeted manner, allowing for a single, generic method that can be deployed across a wide variety of analytes. Another critical advantage of the information-rich data obtained from HRMS systems is that it can be re-investigated at later stages to better understand analyte ADME profiles and screen for metabolites of interest, which can accelerate the progression of drug candidates.
In this work, we describe an analytical workflow used for the extraction of a lipid conjugated antisense oligonucleotide (ASO) from rat plasma using the OligoWorks SPE Microplate Kit (OligoWorks Kit) followed by quantification on a LC-HRMS system. The OligoWorks kit offers a simple, broadly applicable sample preparation solution that provides high recoveries and low matrix effects across a diverse set of therapeutic oligonucleotides from biomatrices. Extracted samples were then subjected to LC-MS analysis using an ACQUITY™ H-Class UPLC™ combined with a Xevo G3 quadrupole-time of flight (QToF) HRMS instrument along with a generic data acquisition method that facilitates sensitive, robust, and efficient quantitative analysis of the analyte demonstrating a standardized analytical workflow that can enhance productivity in discovery bioanalytical laboratories performing therapeutic oligonucleotide analysis.
Experimental
LC-MS Chromatographic Separation and Experimental Conditions
Liquid Chromatography
|
LC system: |
ACQUITY UPLC H-Class PLUS System with FTN |
|
Column: |
ACQUITY Premier Oligonucleotide C18 Column, 130 Å, 1.7 µm, 2.1 x 50 mm |
|
Column temperature (°C): |
55 °C |
|
Sample temperature (°C): |
10 °C |
|
Mobile phase A: |
1% HFIP (Hexafluoro-2-propanol) 0.1% DIPEA (N,N-Diisopropylethylamine) in H2O |
|
Mobile phase B: |
0.75% HFIP (Hexafluoro-2-propanol), 0.0375% DIPEA (N,N-Diisopropylethylamine, 65% ACN 35% H2O |
|
Purge solvent: |
25:25:25:25 Methanol:Acetonitrile:Isopropanol:Water |
|
Injection volume (µL): |
10 µL |
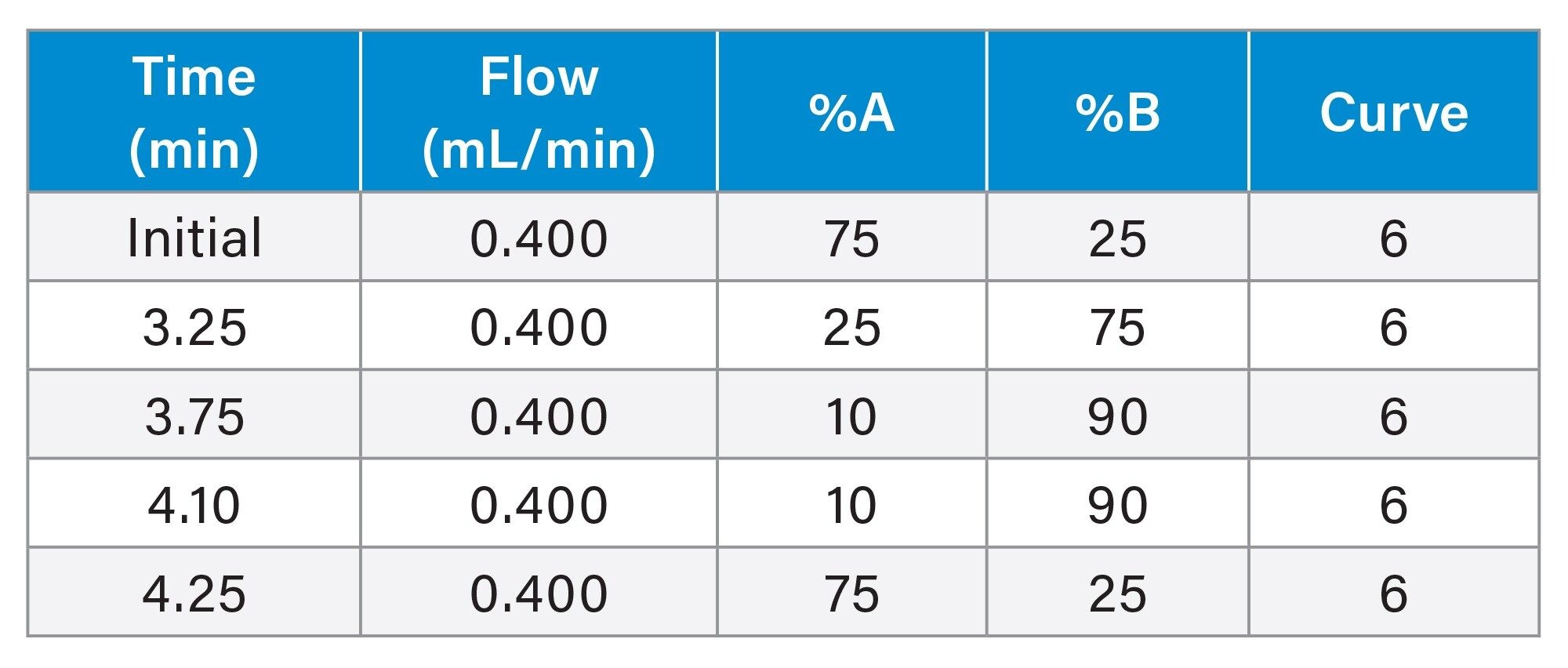
LC Gradient table
MS System Conditions
|
MS system: |
Xevo G3 QToF |
|
Ionization mode: |
ESI- |
|
Acquisition mode: |
MS Full Scan |
|
Capillary voltage (kV): |
2 |
|
Desolvation temperature (°C): |
600 |
|
Desolvation gas flow (L/Hr): |
1000 |
|
Cone gas flow (L/Hr): |
150 |
|
Collision gas flow (L/Hr): |
0.2 |
|
Nebulizer (Bar): |
7 |
Data Management
|
Instrument control software: |
waters_connect™ |
|
Quantification software: |
UNIFI™ |
Chemicals, Reagents, Materials, and Solvents
Lipid conjugated ASO LC-MS standard (Monoisotopic neutral mass 6046.08484 Da) was procured from Waters™ Corporation (Milford, MA USA) (p/n: 186010747). The internal standard (IS) was a custom synthesized lipid conjugated oligonucleotide (Monoisotopic neutral mass 5716.9361 Da) which differed from the analyte of interest by a couple of base pairs, made by BioSearch Tech (Lystrup, Denmark).
MS grade Acetone, Methanol, water, acetonitrile, isopropanol, Hexafluoro-2-propanol (HFIP), N,N-Diisopropylethylamine (DIPEA), and ammonium acetate were purchased from Sigma Aldrich (St. Louis, MO, USA). K2 EDTA ‘Wistar Hannover’ rat plasma was procured from BioIVT (Westbury, NY, USA). DNase/RNase-free distilled water was purchased from ThermoFisher Scientific (p/n: 10977015) and was used for oligonucleotide standard preparation and SPE sample eluate dilution. The OligoWorks Kit (p/n: 186010614) was procured from Waters Corporation (Milford, MA, USA).
OligoWorks SPE Wash Reagent Preparation
OligoWorks SPE Wash 1: 50 mM Ammonium Acetate buffer, pH 5.5 was prepared by weighing out 3.84 g ammonium acetate and bringing to 1 Liter volume and adjusting pH to 5.5.
OligoWorks SPE Wash 2: 2 different wash solutions were evaluated as potential Wash 2 solvents.
- 10% Methanol/90% Water solution was prepared by adding 100 mL of methanol to 900 mL of water
- 30% Methanol/70% Water solution was prepared by adding 300 mL of methanol to 700 mL of water
Stock Solutions, Calibration Curve, and QC Sample Preparation
Lipid conjugated ASO and IS were reconstituted in 50 µL of 20% Acetone:80% DNA/RNAase free water to obtain a stock concentration of 100 nmol/mL for each. Lipid conjugated ASO was spiked in rat plasma to obtain a calibration curve (1–1,000 pmol/mL) and QC points (LQC-5 pmol/mL, MQC-50 pmol/mL and HQC-750 pmol/mL). IS was further diluted 100-fold in 20% Acetone solution to obtain a concentration of 1 nmol/mL. This IS stock was then used to spike in calibration curve and QC points at a final concentration of 50 pmol/mL.
Sample Pretreatment and Extraction Using the Oligoworks SPE Microplate Kit
Triplicates of the prepared calibration curve and QC samples were extracted from rat plasma following the protocol provided in the OligoWorks SPE Kits and Components care and use manual (720008066EN). Briefly, 100 µL of calibration curve and QC samples were added to an Eppendorf 1mL deep well plate and digested using the reagents and protocol supplied in the RapiZyme Proteinase K Digestion Module and subsequently extracted using the OligoWorks SPE Microplate and Eluent.
Results and Discussion
The Lipid Conjugated ASO LC-MS Standard (p/n: 186010747) is a 16 residue gapmer antisense oligonucleotide (ASO) containing a fully phosphorothioated backbone, a 5’ palmitate modification and methoxy ethyl modified termini. Its sequence is shown below. It has a negative control sequence that does not have any homology with mammalian genes. The 5’ modification is created from the use of a 5’-Palmitate-C6 CE phosphoramidite. A chemical structure for this modification is provided below. Asterisks (*) denote PS bonds, MOE stands for methoxy ethyl 2’ modifications, and MeC denotes a 5-methyl cytidine residue.
5’ d 5-Pal-*-MOE-MeC-*-MOE-G-*-MOE-MeC-*C*G*A*T*A*A*G*G*T*A*- MOE-MeC-*-MOE-A-*-MOE-MeC 3’
The internal standard is also a lipid modified antisense oligonucleotide with similar modifications as the analyte and differing from the analyte of interest by only a couple of residues as shown below:
5' d 5-Pal-MOE-MeC-*-MOE-G-*-MOE-MeC-*C*G*A*T*A*A*G*G*T*-MOE-A-*-MOE-MeC-*-MOE-A 3'
OligoWorks SPE Protocol Optimization
Initial plasma recovery and Matrix effects experiments for the lipid conjugated ASO were performed using the starting sample preparation and extraction protocol described in the OligoWorks SPE Care and Use Manual. LC-MS analysis for these experiments were performed using an ACQUITY PREMIER LC System and Xevo TQ-Absolute tandem quadrupole (TQ) instrument and MRM analysis using the [M-9H+]- precursor 671.1 m/z and fragment ion of 95.1 m/z and the same chromatographic conditions and column as described in the methods section. Using the starting protocol resulting recoveries were ≈80% and 60% for the lipid conjugated ASO and IS, respectively. Matrix effects were <15% (Data not shown). Although these recovery values are acceptable, we attempted to further optimize the SPE protocol to enhance the recoveries and maximize sensitivity.
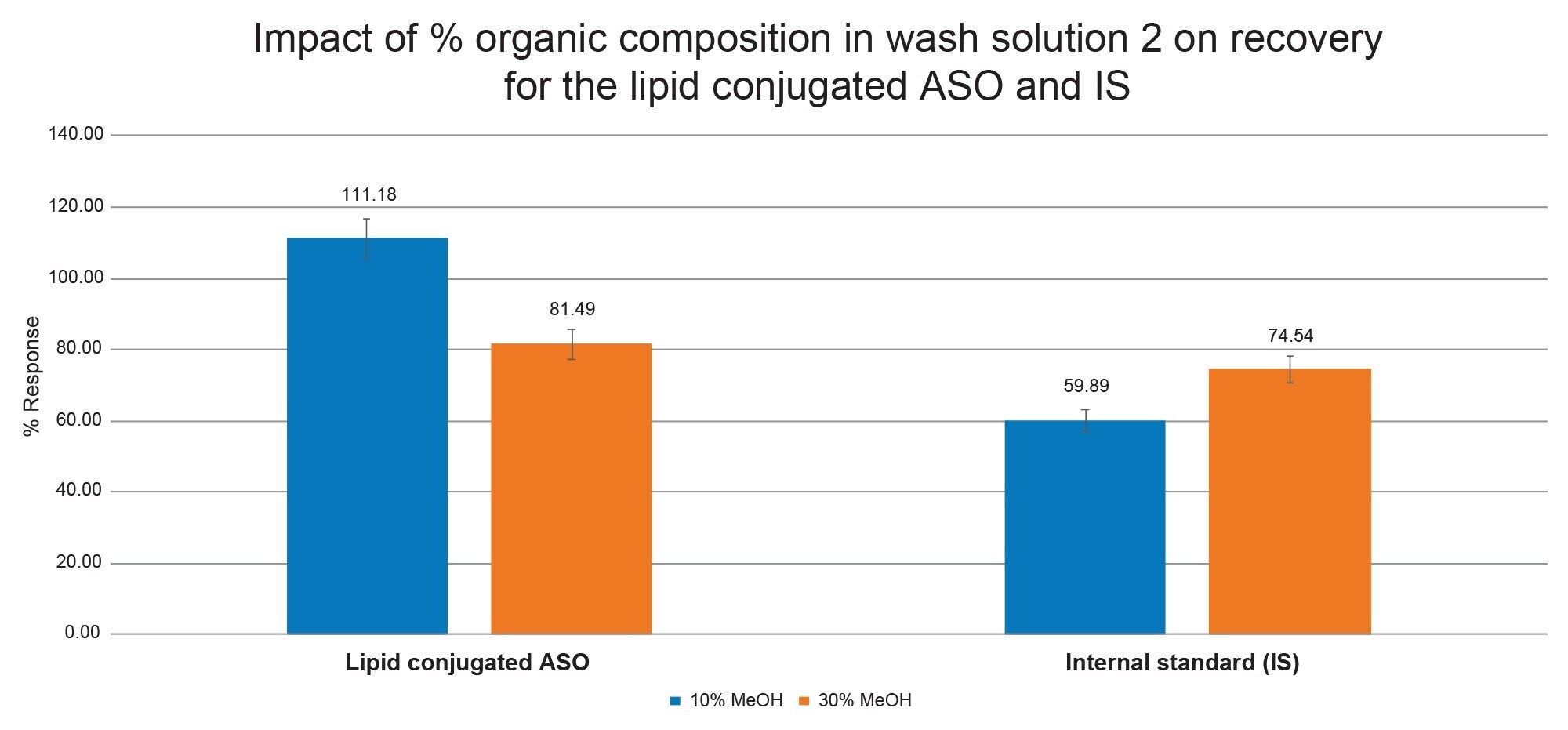
The OligoWorks SPE WAX sorbent is a polymeric reversed-phase, weak anion exchange mixed-mode sorbent. This bi-modal functionality can be leveraged for protocol optimization in the load, wash and elution steps. We started with optimizing the Wash 2 steps as this step can affect retention of the analyte of interest while also impacting removal of unwanted matrix components. A reduction in the organic composition of Wash 2 from 30% methanol (recommended Wash 2 composition in the Care & Use manual) to 10% greatly improved recovery for the target lipid conjugated ASO, but decreased recovery for the IS. In this specific instance, the target lipid conjugated ASO, was partially eluting in the Wash 2 step due to the increase in the methanol composition. In contrast, lower recoveries were seen for the IS when Wash 2 methanol composition was reduced. This performance is shown in Figure 1. As evident from this data, analytes closely related in sequence can still show differential recoveries based on percent organic solvent in Wash 2 step of the SPE.
LC-MS Method Development
LC Gradient Optimization
The LC gradient provided in the OligoWorks SPE care & use manual was used as a starting point for analysis of the lipid conjugated ASO and IS. Using this gradient, the analyte and IS eluted towards the end of the chromatographic run (data not shown). This is likely due to the additional hydrophobicity imparted to the analyte due to the conjugated lipid. The LC gradient was therefore optimized to have starting conditions with a higher % organic solvent and a steeper gradient, going from 25% to 75% B in 3.25 minutes. Using these LC conditions, the analyte and IS peaks eluted in the center of the gradient (data not shown). Additionally, the use of ACQUITY™ Premier Oligonucleotide BEH™ C18 Column minimized non-specific anionic interactions of the oligonucleotides with the column hardware (peak width at base ≈5 seconds) and helped achieve greater sensitivity and reproducibility as these columns are QC tested with oligonucleotides to ensure optimal performance.
HRMS Data Acquisition and Processing
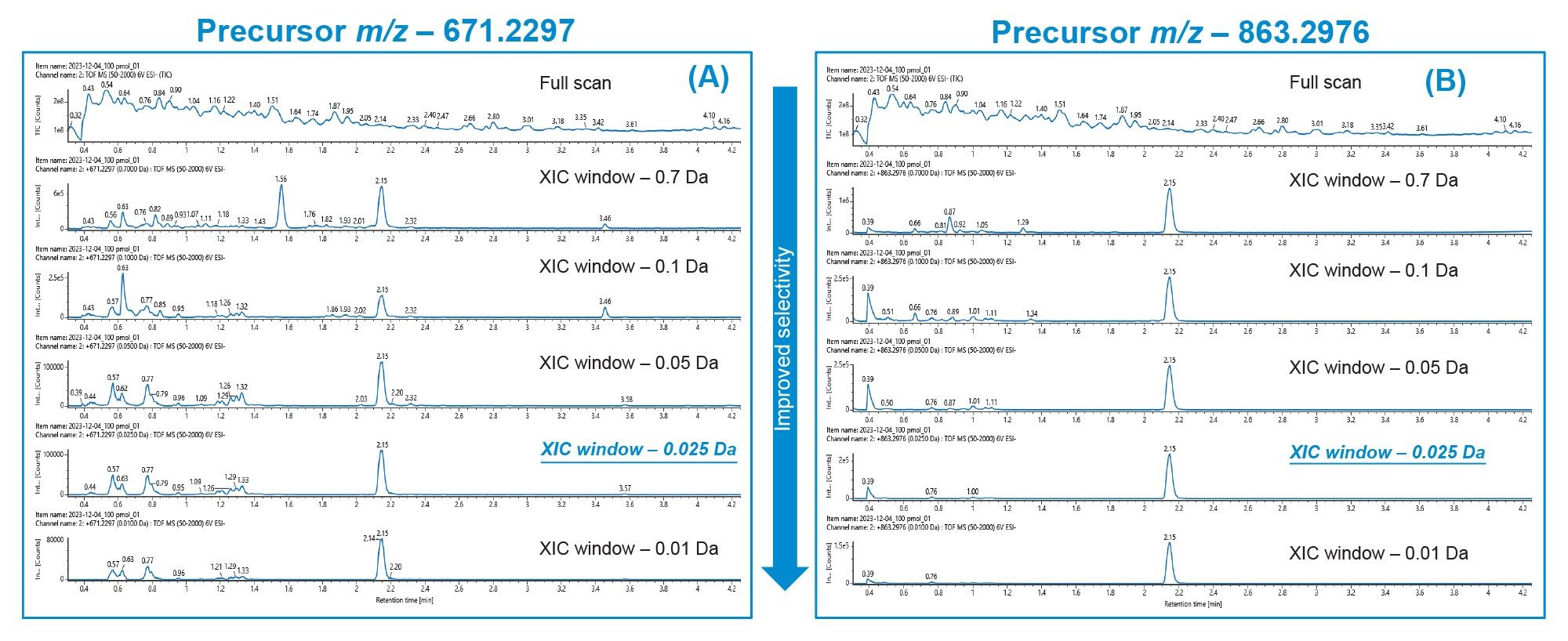
The Xevo G3 QToF MS maximizes sample information for analytes of interest, from detailed characterization to accurate quantitation. A full MS scan experiment (m/z 50–2000, scan time 0.2 sec) was set up using the UNIFI application in waters_connect to collect data in an untargeted manner. This approach allows for a single acquisition method that can be used across a diverse set of analytes without the need for added method development. Figure 2 shows the MS spectra for precursor [M-9H+]- at m/z 671.2297 (Figure 2A) and precursor [M-7H+] at m/z 863.2976 (Figure 2B) for a 100 pmol/mL standard of the lipid conjugated ASO extracted from rat plasma.
![Representative mass spectra of Lipid conjugated ASO LC-MS standard precursors at (A) [M-9H+]- (m/z 671.2297) and (B) [M-7H+]- (m/z 863.2976)](/content/dam/waters/en/app-notes/2024/720008256/720008256en-f2.jpg.82.resize/img.jpg)
In addition to the ease of data acquisition, the high resolution of the Xevo G3 QToF allows for a deeper investigation of the data and enables the use of narrow mass extraction (XIC) windows to achieve maximum signal-to-noise (S/N). A TQ instrument is routinely operated at unit resolution of +/- 0.7 Da. The high resolving power of the HRMS system can consistently differentiate masses separated by m/z of 0.001 Da. While processing HRMS data, different XIC windows can be evaluated to eliminate MS signal from unwanted matrix components, while focusing on the m/z of the analyte of interest to ensure sufficient signal. This allows the users to minimize the background noise while maintaining the analyte signal, thereby increasing S/N. However, after a certain point, tightening the XIC window starts to negatively impact the analyte peak area counts, without having a significant impact on the background peaks in the chromatogram. Finding the ideal XIC window is therefore a balance between minimizing signal from unwanted background components without compromising the analyte peak area counts.
For the lipid conjugated ASO, XIC windows of 0.7 to 0.01 Da were evaluated. As evident from Figure 3, as the XIC window narrows the non-specific background peaks from matrix components continue to get smaller, providing a cleaner chromatographic background and improving assay selectivity. For final quantification, we chose a XIC window of 0.025 Da as it gave the best balance between a clean background and sufficient analyte peak area counts.
Sensitive, accurate, and precise
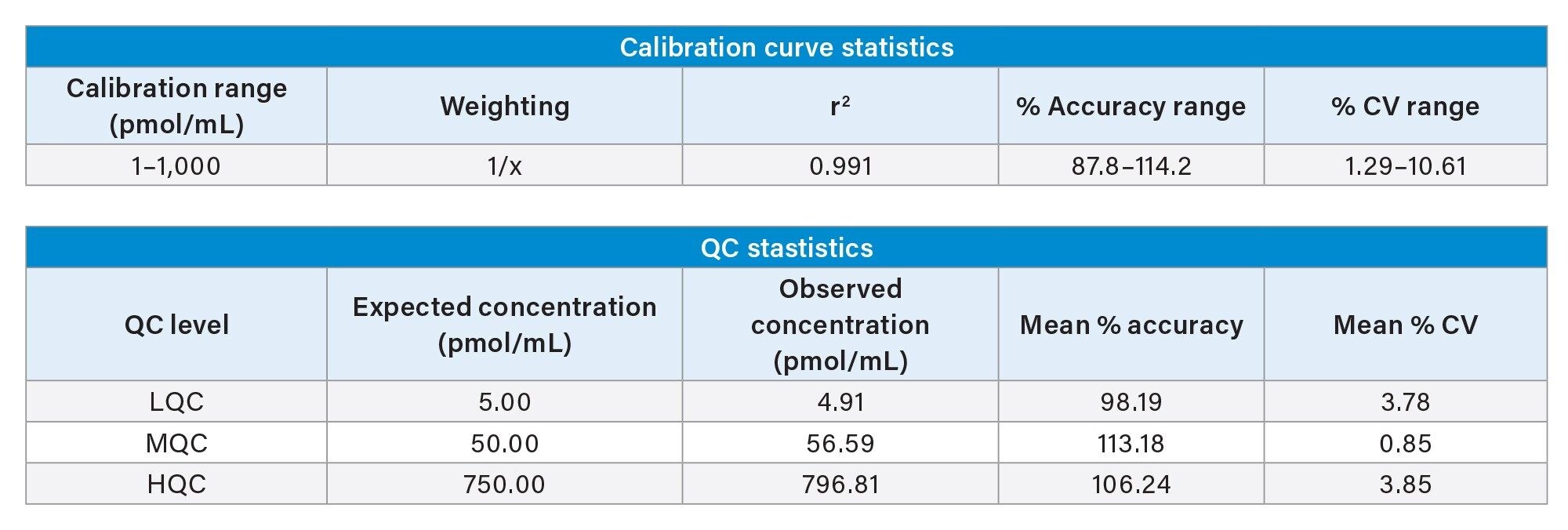
The lipid conjugated ASO spiked in rat plasma, extracted using the OligoWorks Kit and analyzed on the Xevo G3 QToF MS system was linear from 1–1,000 pmol/mL, with a linear fit (r2) of 0.991 using a 1/x weighting. The % accuracy range across all points on the calibration curve was 87.8–114.2 and % CV range across all points on the curve was 1.29–10.61. (Table 1)
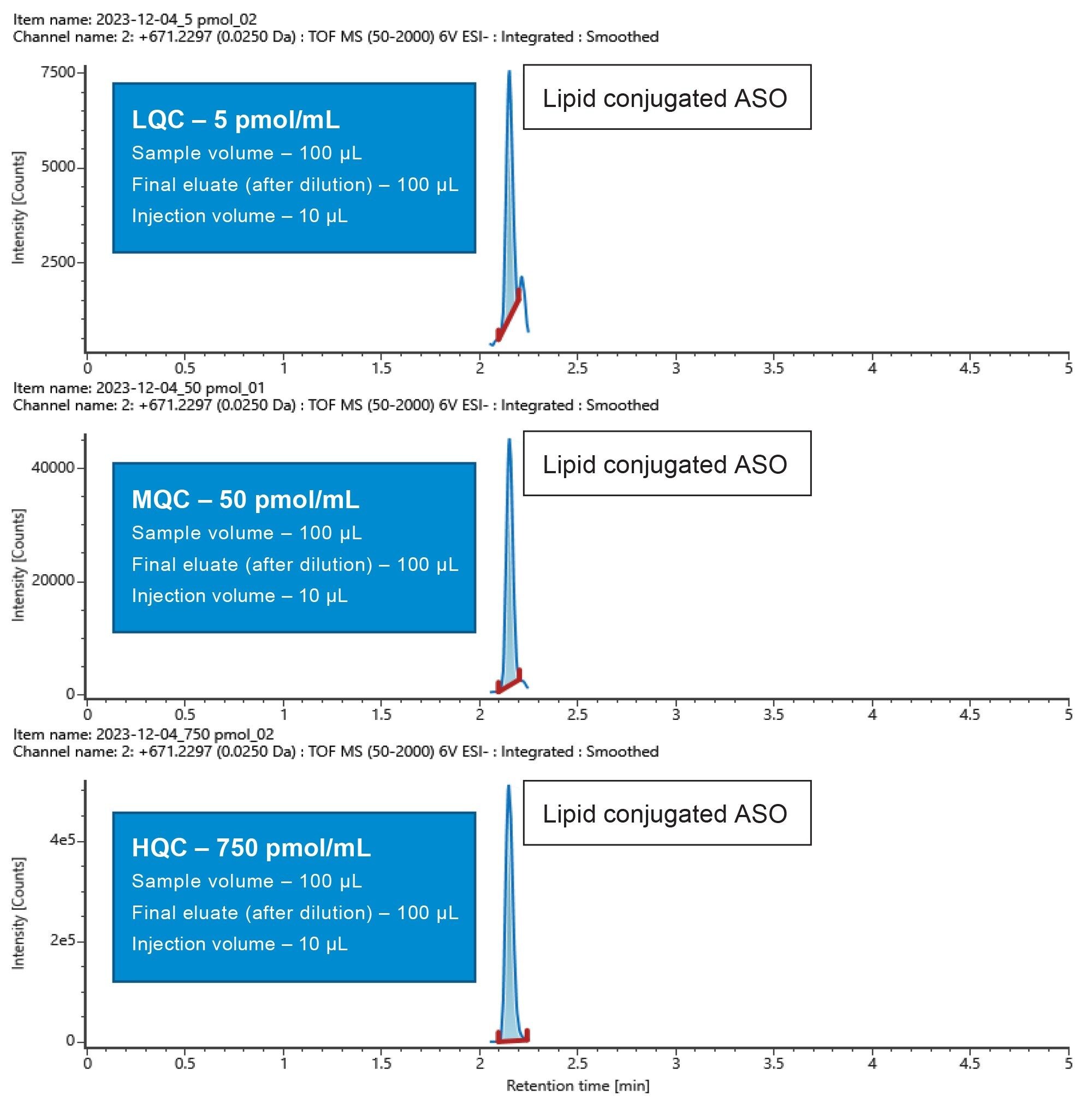
The mean % accuracy and mean % CV for the LQC (5 pmol/mL), MQC (50 pmol/mL), and HQC (750 pmol/mL) were 98.19%, and 3.78%, 113.18%, and 0.85%, and 106.24%, and 3.85% respectively (Table 1). The area counts for the calibration curve and QC points increased linearly with an increase in concentration, as highlighted in the representative QC chromatograms in Figure 4.
Conclusion
Discovery bioanalysis laboratories are often looking for standardized, streamlined workflows to support the large number of projects and analytes they are required to support. To facilitate greater efficiency and productivity in this regard, we have demonstrated a simple, easy-to-implement workflow using the OligoWorks SPE Microplate kit for extracting the analyte of interest from biological matrix, followed by untargeted quantification of the analyte on the Xevo G3 QToF HRMS instrument.
For the lipid conjugated ASO LC-MS standard, we were able to achieve a linear calibration curve from 1–1000 pmol/mL, with all calibration curve and QC points passing the FDA bioanalytical method validation guidelines.
720008256, February 2024