Comprehending COVID-19: Distinct Identification of the SARS-CoV-2 Delta Variant Using the SARS-CoV-2 LC-MS Kit (RUO)
For research use only. Not for use in diagnostic procedures.
This is an Application Brief and does not contain a detailed Experimental section.
Abstract
As of July 2021, the SARS-CoV-2 Delta (B.1.617.2) variant is rapidly becoming the dominant form of the virus across the globe. The SARS-CoV-2 LC-MS Kit (RUO) was developed to detect signature peptides associated with the wildtype SARS-CoV-2 NCAP protein. Using the GISAID database to track on-going mutations, we have established that tryptic peptides generated from Delta variant can still be captured and quantified using SARS-CoV-2 LC-MS Kit (RUO) SISCAPA antibody. In addition, the Delta variant can be selectively identified and differentiated from the wildtype peptide found in the other variants of concern using chromatographic separation and Multiple Reaction Monitoring (MRM).
Benefits
- SARS-CoV-2 Delta variant can be identified using the SARS-CoV-2 LC-MS Kit (RUO)
- SARS-CoV-2 Delta variant can be differentiated from other variants of concern using the SARS-CoV-2 LC-MS Kit (RUO)
Introduction
In the rapidly changing landscape of the COVID-19 pandemic, the requirement to adapt and respond to changes in the SARS-CoV-2 virus has never been more important. As of July 2021, the SARS-CoV-2 Delta (B.1.617.2) variant is rapidly becoming the dominant form of the virus across the globe. The mRNA and translated amino acid sequence of SARS-CoV-2 are used as targets for many analytical technologies for identifying presence of the virus. The Delta variant contains mutations that could potentially impact these target sites and thereby affect the suitability of these platforms for identifying SARS-CoV-2.
The SARS-CoV-2 LC-MS Kit (RUO) was released to aid the direct analysis of three SARS-CoV-2 Nucleocapsid (NCAP) Peptides; AYNVTQAFGR (AYN), ADETQALPQR (ADE), and NPANNAAIVLQLPQGTTLPK (NPA) in clinical research studies. It has been demonstrated that 3 amol/µL of each peptide in viral transport medium can be detected using Stable Isotope Standards with Capture by Anti-Peptide Antibodies (SISCAPA) and the ACQUITY UPLC I-Class and Xevo TQ-XS Mass Spectrometer.1
The impact of the variants on these target peptides found in the Kit have been monitored through the use of the GISAID database.2 Data on previous variants of concern; Alpha, Beta, and Gamma, were extracted and examined and there were no mutations noted that impact the target peptide sequences. However, upon examining the Delta variant mutations, it was noted that the ADE peptide was affected, resulting in amino acid substitution adjacent to the N-terminus (D377Y) and a less prevalent substitution at the C- terminus (R385K) of the peptide (AYETQALPQK).
This application brief demonstrates the Delta variant mutations can still be captured using SARS-CoV-2 LC-MS Kit (RUO) SISCAPA Ab and be selectively identified and differentiated from the wildtype peptide found in the other variants of concern using chromatographic separation and Multiple Reaction Monitoring (MRM).
Results and Discussion
The SARS-CoV-2 LC-MS Kit (RUO) was used to perform the studies detailed in this application note, following the Instructions for Use (IFU).
The Kit workflow describes the denaturation, digestion, enrichment, and UPLC-MS/MS analysis of three peptides; AYNVTQAFGR (AYN), NPANNAAIVLQLPQGTTLPK (NPA), and ADETQALPQR (ADE) derived from the SARS-CoV-2 NCAP protein.
The peptides AYETQALPQR, ADETQALPQK and AYETQALPQK were synthesized (Lifetein, NJ, USA) and added to a solution containing ADETQALPQR (ADE), AYNVTQAFGR (AYN), and NPANNAAIVLQLPQGTTLPK (NPA). The peptides were added to viral transport medium and extracted in replicates using the SARS-CoV-2 LC-MS Kit (RUO) and analyzed over a single experimental run.
Skyline (MacCoss Lab Software, WA, USA) was used to identify potential MRM transitions for the variant peptides and analysis was performed using the ACQUITY UPLC I-Class and Xevo TQ-XS Mass Spectrometer.
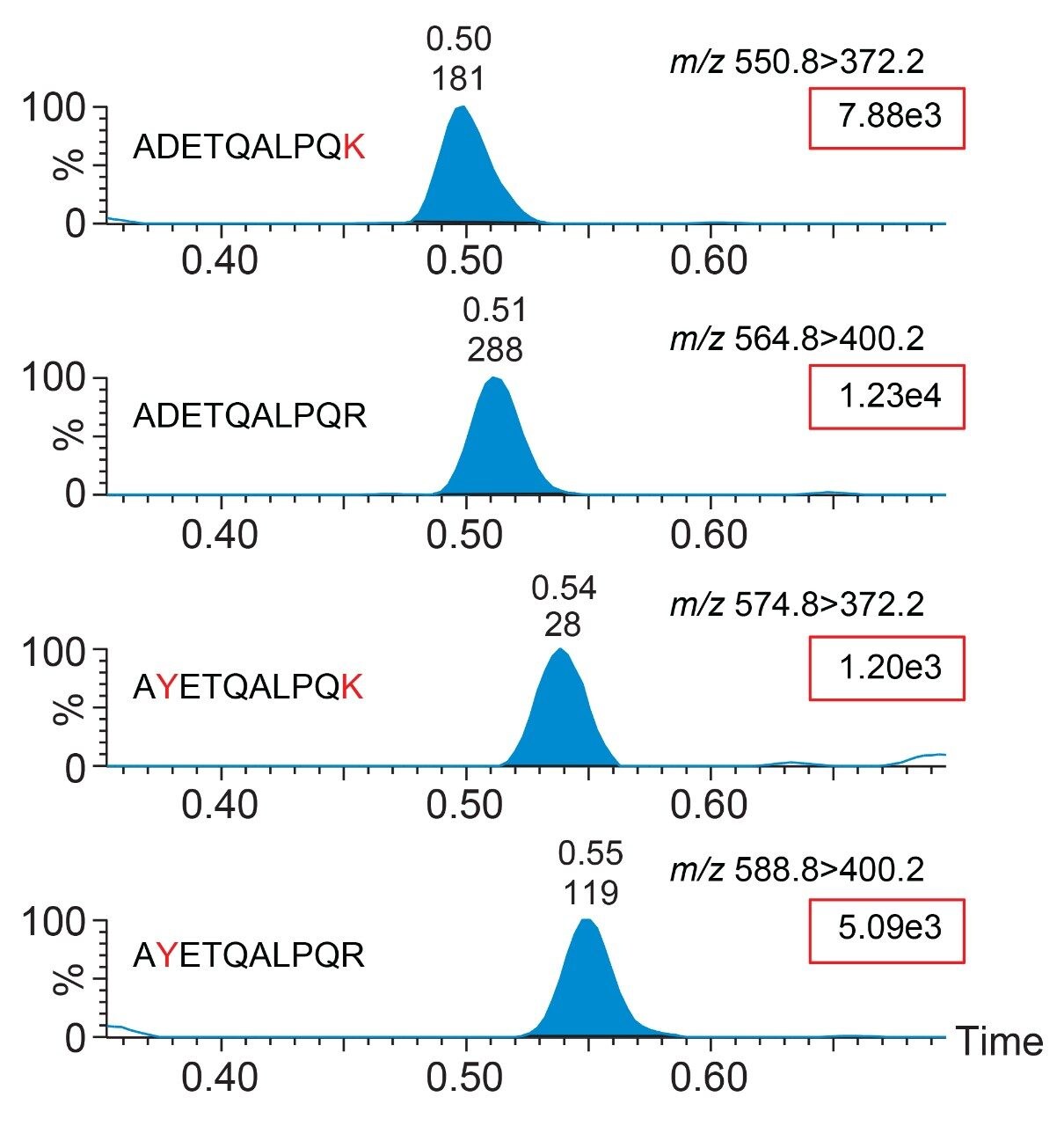
The peptide variants can be identified through the use of specific MRM transitions, with small deviations in retention time noted between the peptides (Figure 1A). No significant changes to the method are required in order to identify these new peptide targets.
Performed over a single experimental run, all variant peptides are captured by the ADE wildtype monoclonal antibody using SISCAPA. However, it is noted that the double mutation (D377Y/R385K) causes significant decrease in analytical sensitivity of the method relative to the other peptides (Figure 1B) but the detection remains reproducible across five replicates for all spiked peptides (≤8.9% RSD). The D377Y/R385K variant peptide is still identifiable at concentrations >10 amol/µL (Figure 2).
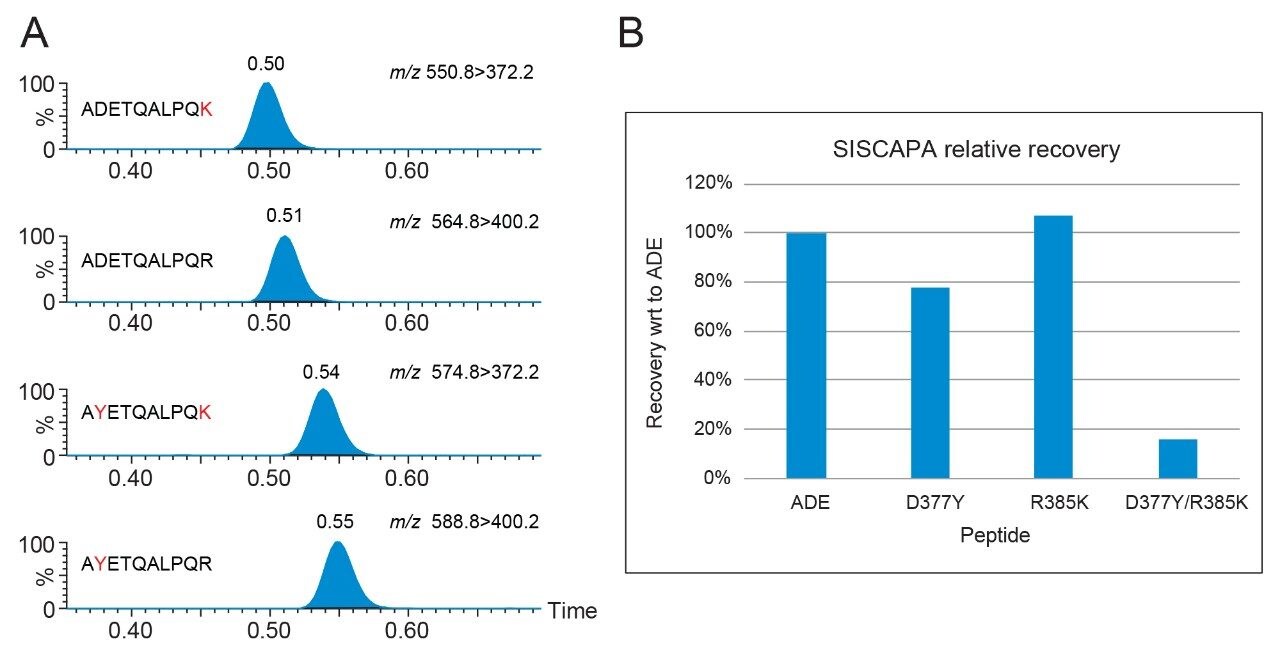
Figure 1B. SISCAPA relative recovery (n=5 for each peptide) of ADETQALPQR, AYETQALPQR (D377Y), ADETQALPQK (R385K), and AYETQALPQK (D377Y/R385K).
Conclusion
Using the Waters SARS-CoV-2 LC-MS Kit (RUO) we were able to successfully capture and identify peptide variants associated with the SARS-CoV-2 Delta (B.1.617.2) variant.
The D377Y substitution is the most prevalent mutation when analyzing the GISAID database from March 2020 to July 2021 (55%), with the R385K and the D377Y/R385K double substitution prevalence estimated to be less than 0.045%. Based on a single experimental run, the Delta variant peptides and the peptides associated with other variants of concern (Alpha, Beta, and Gamma) can be differentiated by chromatographic separation and simultaneous monitoring of MRM transitions. Therefore, it has been established that the SARS-CoV-2 LC-MS Kit (RUO) is suitable for the LC-MS/MS analysis of the predominant SARS-CoV-2 Delta variant NCAP peptides for clinical research activities.
References
- Foley D, Wardle R, Ferries S, Pattison R, Warren J, Calton L. Advancing Research with the SARS-CoV-2 LC-MS Kit (RUO). Waters Application Note 720007266, May 2021.
- GISAID database. https://www.gisaid.org/hcov19-variants/.
720007353, August 2021