
For research use only. Not for use in diagnostic procedures.
This is an Application Brief and does not contain a detailed Experimental section.
This application brief demonstrates the workflow to automate data processing in TargetLynx and Skyline for MetaboQuan-R and LipidQuan targeted assays. The pipelines are configured to automatically generate a results report, which is directly compatible with MetaboAnalyst.
Increase throughput of MetaboQuan-R and LipidQuan assays with automation of the data processing workflow in TargetLynx and Skyline using Symphony.
MetaboQuan-R and LipidQuan are two targeted assays designed to compare samples from large cohorts and highlight potential markers of interest in a rapid, high-throughput manner. All LC and MS methods for both assays are provided in a Quanpedia database and do not require any further development. More specifically, MetaboQuan-R allows for the rapid screening and semi-quantitative analysis of different compound classes (acylcarnitines, bile acids, amino acids, triglycerides, and peptides) using the same LC conditions. LipidQuan is a comprehensive and high-throughput HILIC-based methodology for the separation and quantification of polar and non-polar lipid classes. Here, we show how the use of the Symphony Software improves the efficiency of data processing and thereby increases the throughput of these assays.
The Symphony data pipeline is specifically designed to form custom chains of data processing steps to improve the efficiency in complex LC-MS analyses. The MetaboQuan-R and LipidQuan generated data can be processed using TargetLynx and/or Skyline (MacCoss Lab, University of Washington). The outputs from these processed datasets can be visualized and statistically analyzed with the freely available MetaboAnalyst software to determine potential markers of interest. In this technology brief, we demonstrate how Symphony can be applied to streamline the processing and automatically generate a report compatible with MetaboAnalyst from TargetLynx and/or Skyline.
The pipeline created in Symphony to generate a report from TargetLynx is described in Figure 1. Data are acquired using MassLynx prior to being loaded into TargetLynx. The Symphony pipeline triggers the transfer of the raw data to a processing computer and/or server for storage. The processing is then automated to generate an .xml output from TargetLynx and to convert the file, via an Excel macro, to a .csv results file. The resulting .csv file is compatible for import into MetaboAnalyst, allowing additional interrogation and statistical analysis.
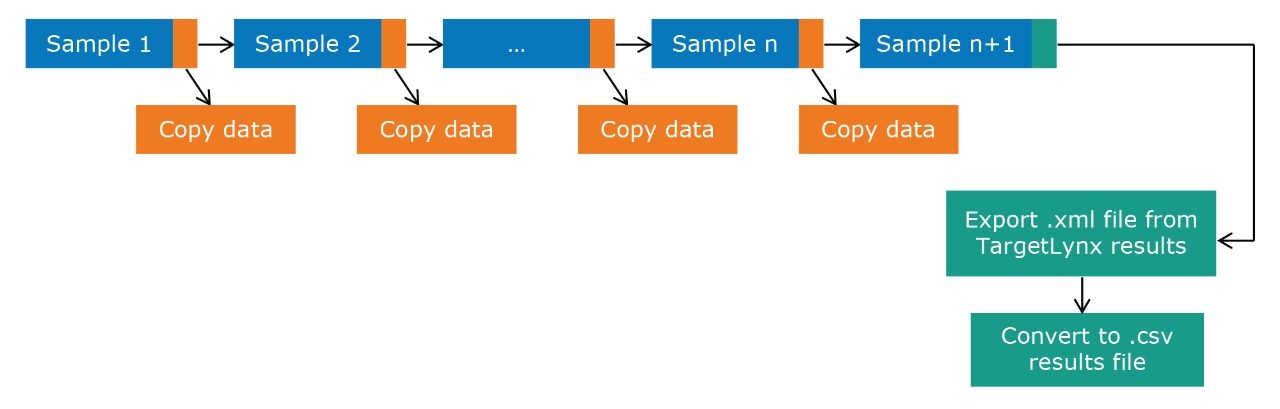
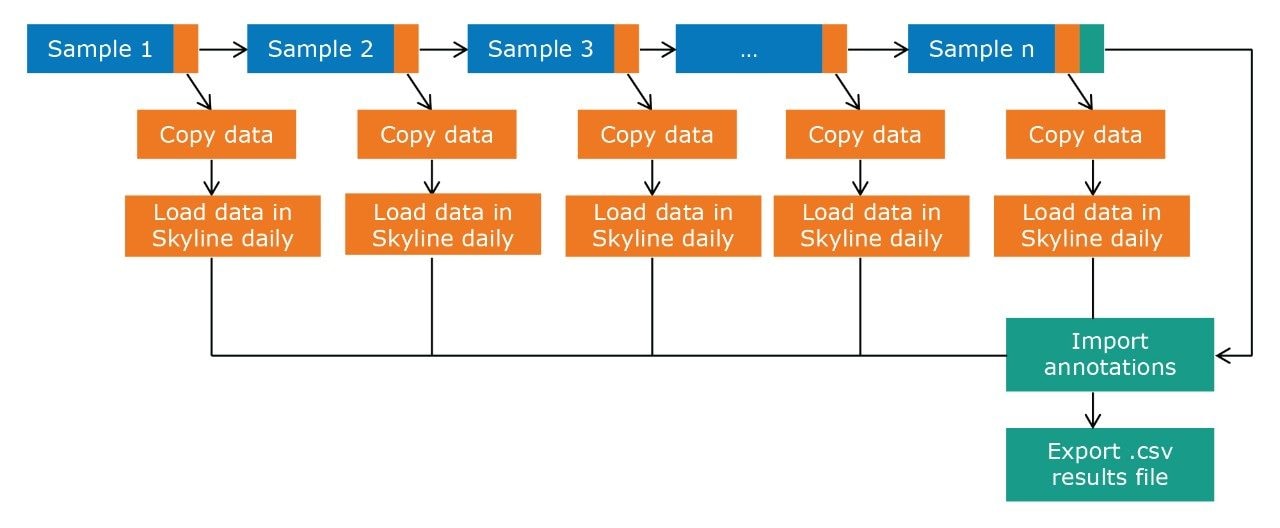
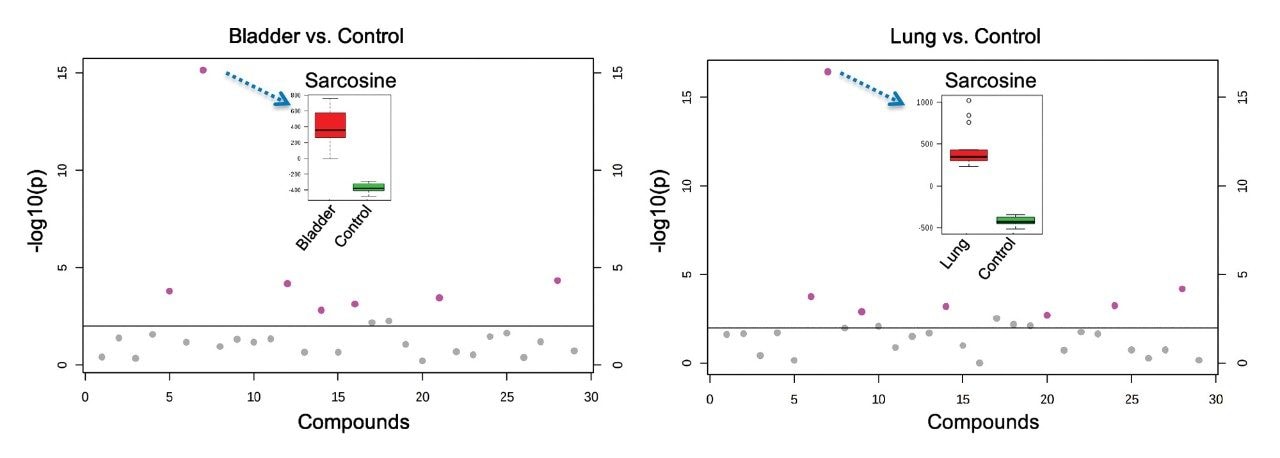
Figure 2 outlines a pipeline created using Symphony for data processing and report generation through Skyline. The pipeline consists of three tasks: Data transfer to a storage server and/or processing PC, data import and processing using Skyline, and finally data export as a .csv result file. On completion of the MassLynx analysis batch, two files are obtained: A Skyline file containing all the raw data and a .csv file consisting of 1) peak area for each sample (raw, normalized against an internal standard or light/heavy ratios) and 2) sample conditions/groups. Key user parameters for each pipeline are shown in Figures 3 and 4. An example amino acid highlighted using the MetaboQuan-R assay and processed via the Skyline/MetaboAnalyst pipeline for a lung and bladder cancer sample set2 is provided in Figure 5.
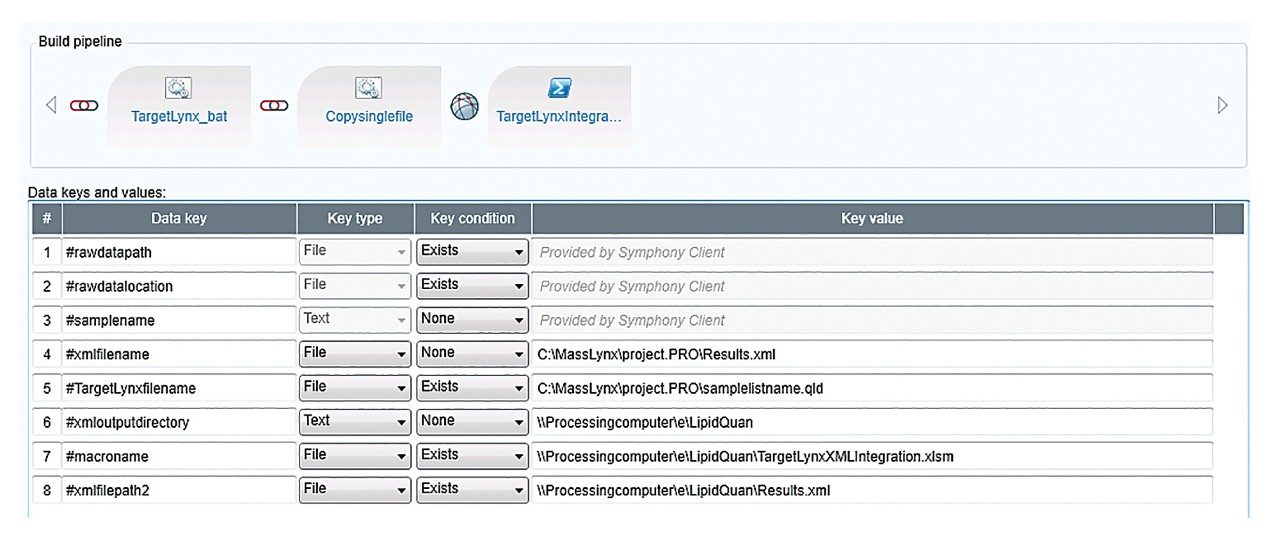
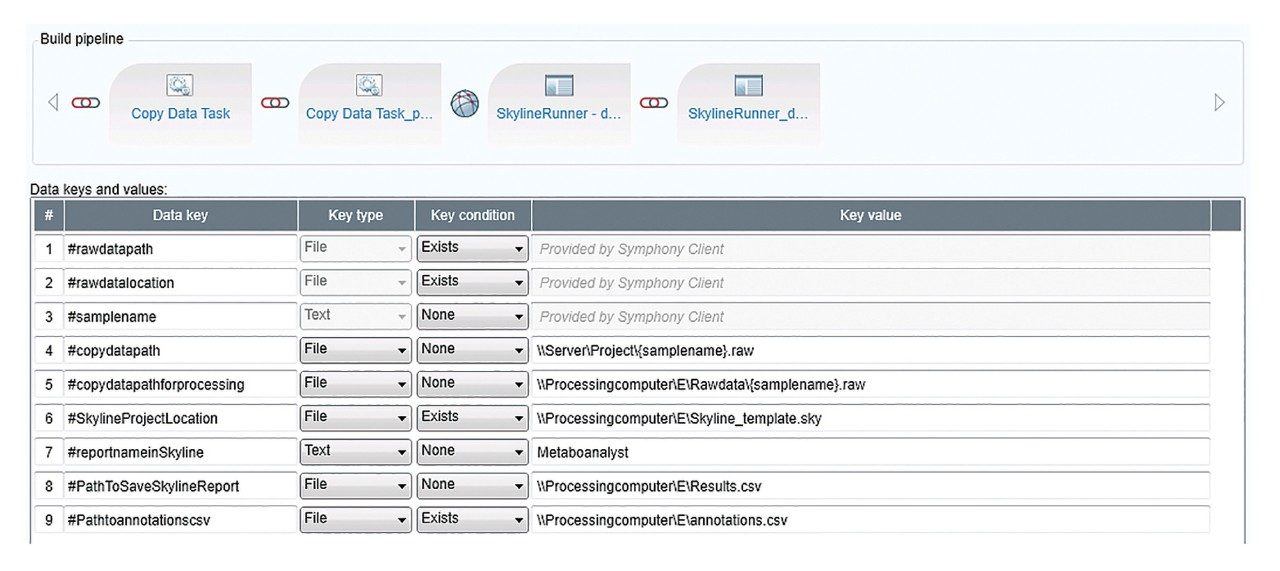
In this application brief, we outlined the application of the Symphony data pipeline software to automate the data processing for the MetaboQuan-R and LipidQuan-based assays. Symphony is triggered from the MassLynx sample list, enabling data transfer prior to loading and processing in TargetLynx and/or Skyline, which generates a report compatible with MetaboAnalyst. Symphony provides the capability of automating data transfer and processing, which ultimately increases the overall workflow productivity and is particularly applicable for large cohort/high-throughput analysis.
720006671, September 2019