
In this application note, chloramphenicol was analyzed with a three-minute runtime using Waters ACQUITY UPLC System, coupled with Xevo TQD and MassLynx Software. The Xevo TQD is a reliable, reproducible, and accessible tandem quadrupole mass spectrometer for routine quantitative and qualitative trace analysis. It incorporates RADAR Technology, which allows for the simultaneous acquisition of multiple reaction monitoring (MRM) transitions and full spectrum data. RADAR was used for method development and background monitoring during the analyses.
Chloramphenicol, an effective broad spectrum antibiotic, is widely used in medicinal and veterinary practices. Its use in humans is restricted due to potential harmful effects. Chloramphenicol is reported to be a cause of a potentially fatal blood condition called idiosyncratic aplastic anemia, and hypersensitivity to the drug affects around one in 30,000 people, regardless of dosage.1 It is also anticipated to be carcinogenic. As a consequence, chloramphenicol is not approved for use in food-producing animals. However, due to its wide availability and low cost, it is used to prevent bacterial infections in aquaculture, apiculture, and poultry farming. Chloramphenicol levels in animal products are strictly monitored. In Europe, the minimum required performance limit (MRPL) for chloramphenicol is 0.3 µg/kg in any food of animal origin,2 and similar limits have been adopted in other countries, including the United States.
In this application note, chloramphenicol was analyzed with a three-minute runtime using Waters ACQUITY UPLC System, coupled with Xevo TQD and MassLynx Software. The Xevo TQD is a reliable, reproducible, and accessible tandem quadrupole mass spectrometer for routine quantitative and qualitative trace analysis. It incorporates RADAR Technology, which allows for the simultaneous acquisition of multiple reaction monitoring (MRM) transitions and full spectrum data. RADAR was used for method development and background monitoring during the analyses.
|
UPLC system: |
ACQUITY UPLC |
|
Runtime: |
3.0 min |
|
Column: |
ACQUITY UPLC BEH C18 1.7 μm, 2.1 x 50 mm |
|
Column temp.: |
55 °C |
|
Sample temp.: |
4 °C |
|
Mobile phase A: |
Water |
|
Mobile phase B: |
Methanol |
|
Weak wash: |
1:1 water:acetonitrile |
|
Strong wash: |
Acetonitrile |
|
Flow rate: |
0.5 mL/min |
|
Injection volume: |
10 μL |
|
Time (min) |
Flow rate (mL/min) |
%A |
%B |
Curve |
|---|---|---|---|---|
|
Initial |
0.5 |
95 |
5 |
N/A |
|
0.4 |
0.5 |
95 |
5 |
6 |
|
1 |
0.5 |
0 |
100 |
6 |
|
1.5 |
0.5 |
0 |
100 |
6 |
|
1.55 |
0.5 |
95 |
5 |
6 |
|
3 |
0.5 |
95 |
5 |
6 |
|
MS system: |
Xevo TQD |
|
Ionization mode: |
ESI negative |
|
Capillary voltage: |
1.0 kV |
|
Source temp.: |
150 °C |
|
Desolvation temp.: |
500 °C |
|
Desolvation gas: |
1000 L/hr |
|
RADAR method: |
(see Figure 1) |
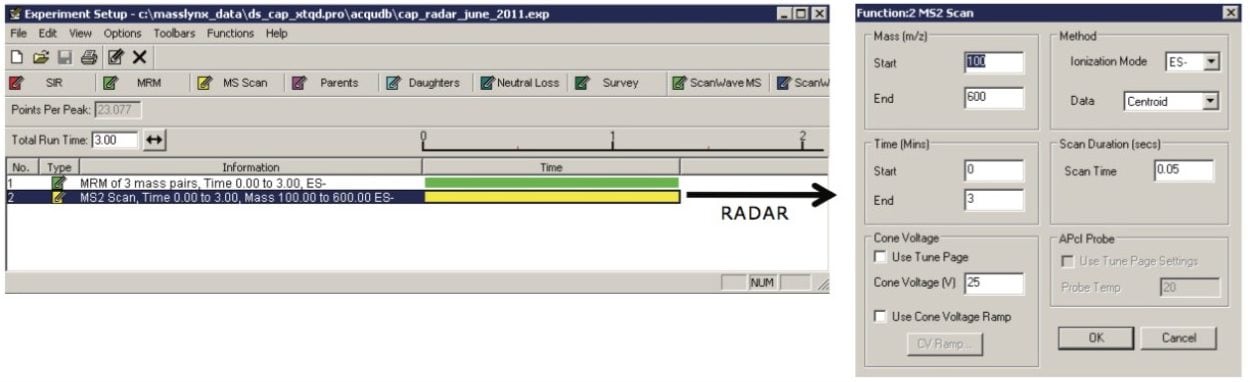
|
MS2 full scan range: |
100 to 600 amu |
|
Scan time: |
0.05 sec |
|
Scan speed: |
10,000 amu/s |
Chloramphenicol (CAS# 56-75-7) was purchased from Sigma-Aldrich. A 1-mg/mL solution of chloramphenicol-d5 in methanol, purchased from Cambridge Isotope Laboratories, was used as an internal standard. Each working solution was prepared at 100 ng/mL in methanol. The calibration curve was prepared with different concentrations of chloramphenicol standard ranging from 0.5 to 10.0 ng/mL, and a fixed amount of chloramphenicol-d5 at 5.0 ng/mL in water.
Sample preparation followed the protocol described by Xi Xia et al. 20103 with minor modifications. The homogenized chicken was spiked with the internal standard, and extraction was performed with ethyl acetate. The supernatant was evaporated to dryness. The residue was re-dissolved in methanol then mixed with 10 mL 4% NaCl solution. Hexane was added and the resulting mixture was vortexed and centrifuged. The upper hexane layer was then discarded and the lower layer subjected to SPE cleanup.
For SPE, an Oasis HLB (3 cc) Cartridge was preconditioned sequentially with 2 mL methanol and 2 mL water. The sample extract was loaded onto the cartridge and passed under vacuum. The cartridge was then rinsed with 3 mL water, followed by 2 mL of 20% methanol. The compounds were eluted from the cartridge using 4 mL of methanol. The eluate was evaporated to dryness at 40 °C under a stream of nitrogen, and the residue was reconstituted in 0.5 mL of water. This solution was filtered through a Waters 0.2 µm PTFE filter prior to UPLC-MS/MS analysis.
The data were acquired and processed using MassLynx 4.1. Software with TargetLynx Application Manager.
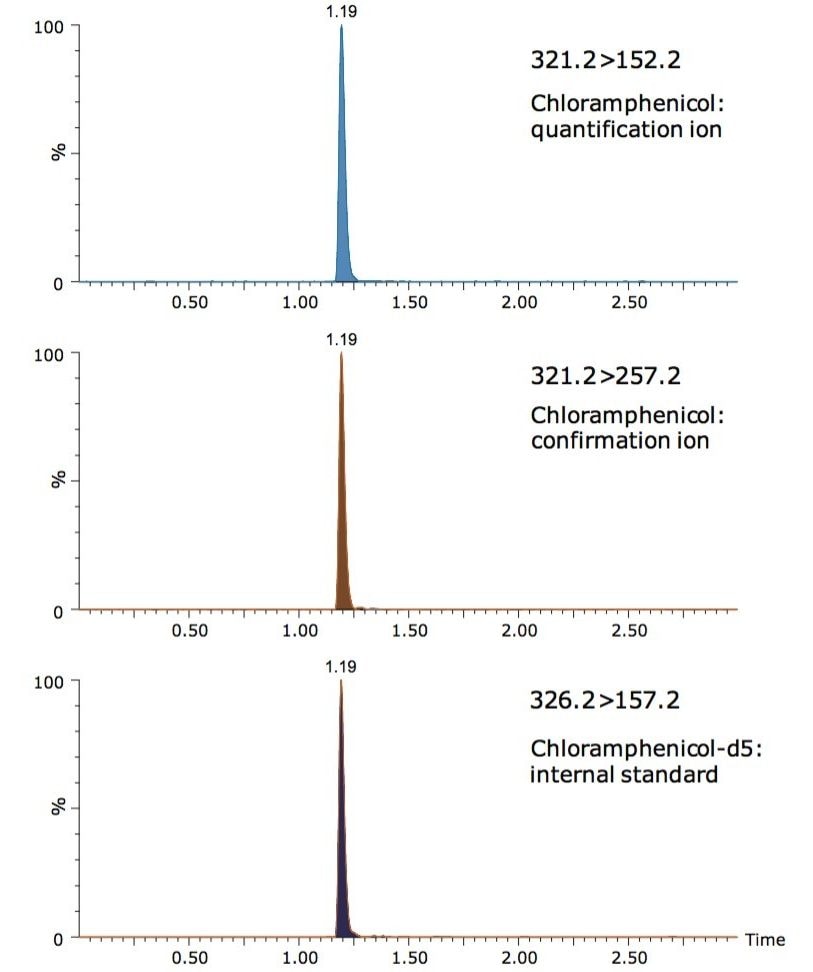
Solutions of chloramphenicol and the internal standard at 1 µg/mL in 50% methanol were used to obtain tuning parameters with IntelliStart Technology. IntelliStart greatly simplifies the use of LC-MS systems by automating instrument setup, compound tuning, and performing system suitability checks. The m/z of both the analyte and internal standard, as well as the cone voltages resulting from this automated tuning are shown in Table 2. The resulting MRM chromatograms from a three-minute UPLC separation of chloramphenicol at 3 ng/mL (equivalent to 0.3 µg/kg in chicken), and the internal standard at 5 ng/mL are shown in Figure 2.

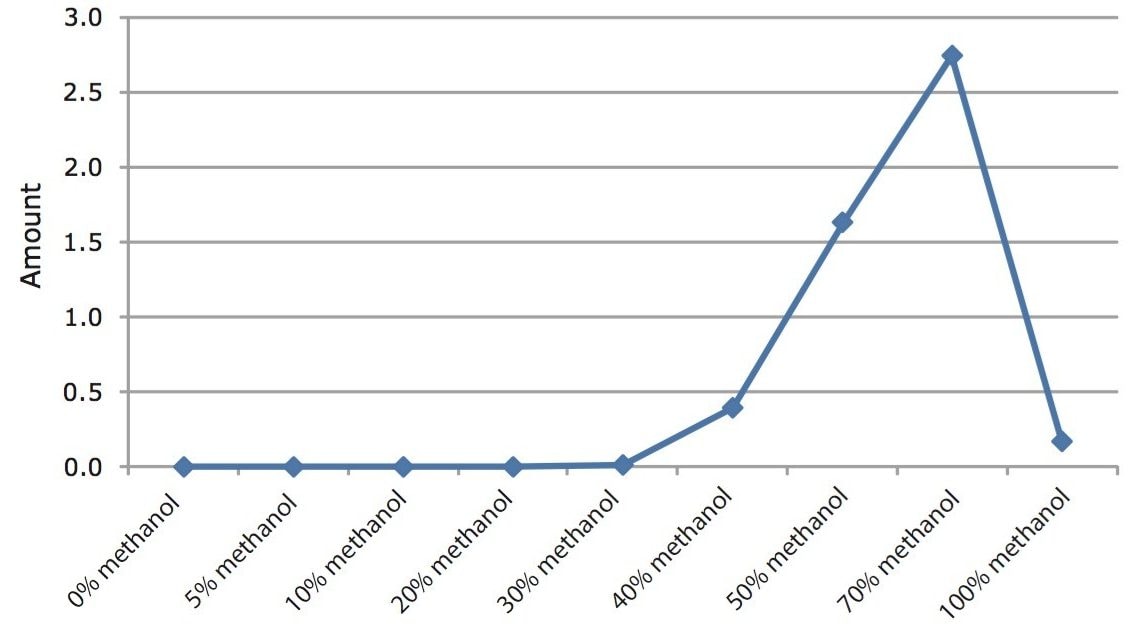
The SPE protocol described by Xi Xia et al.3 was optimized using a solution of chloramphenicol in water at 3 ng/mL. Following loading on to the Oasis HLB Cartridge, the cartridge was washed successively with 2 mL of 5%, 10%, 20%, 30%, 40%, 50%, 70%, and 100% methanol in water. The elution profile of chloramphenicol from the cartridge is shown in Figure 3. Following the 30% wash step, chloramphenicol started to elute from the cartridge. A wash of 20% methanol was selected to prevent any breakthrough of the analyte. To ensure complete elution of chloramphenicol from the cartridge, 4 mL of 100% methanol was chosen for elution.
The best choice for a wash solvent in SPE is one that removes as many matrix interferences as possible without eluting the analyte. In this study, 20% methanol was selected as the wash. Using RADAR Technology, which provides the simultaneous acquisition of full scan and MRM transitions in one analysis, the impact of the selection of the weaker wash on the background matrix was monitored. In Figure 4, the BPI chromatograms from a spiked chicken breast sample following SPE with a 20% methanol wash and a 50% methanol wash are shown. From these data it can be seen that the 50% wash removes more interferences, but at the expense of the analyte, as shown in Figure 3. RADAR Technology can be further utilized in the method development to assess whether any matrix components interfere with the quantification of the analyte, as described below.
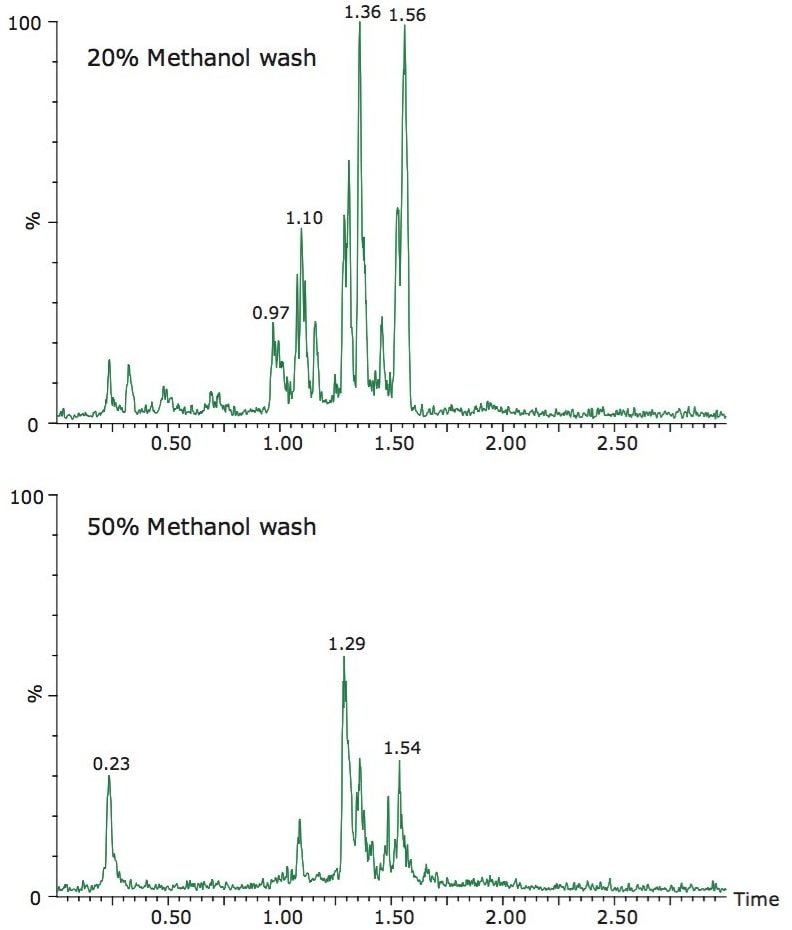
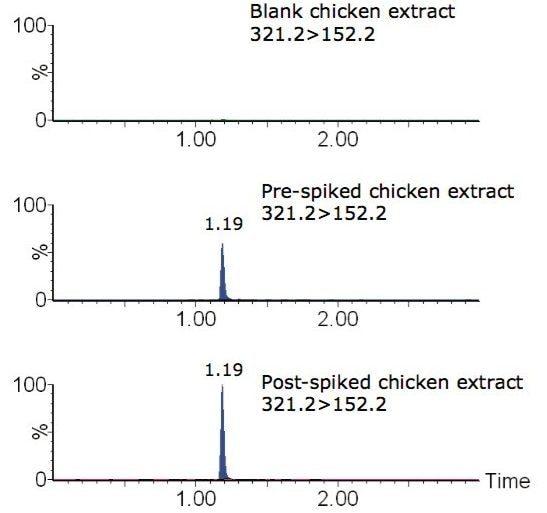
Figure 5 shows MRM chromatograms of a blank chicken extract, chicken extract that was spiked with chloramphenicol prior to extraction (pre-spiked), and chicken extract that was spiked with chloramphenicol following SPE (post-spiked) equivalent to 0.3 µg/kg in tissue, i.e. at the MPRL.
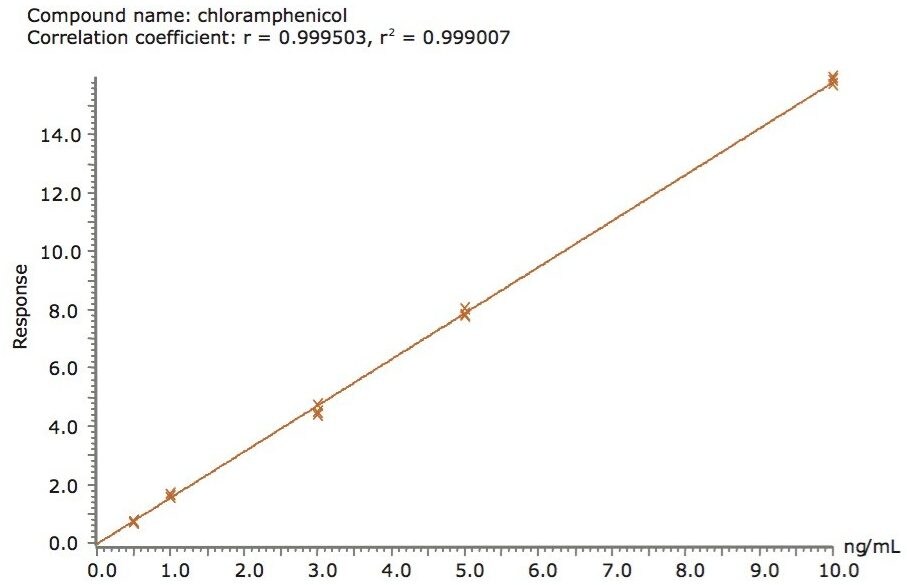
To quantify the chloramphenicol in chicken, calibration solutions were injected in triplicate. The resulting calibration showed excellent linearity across the range of concentrations with a correlation coefficient (r2) of 0.999. An example is shown in Figure 6.
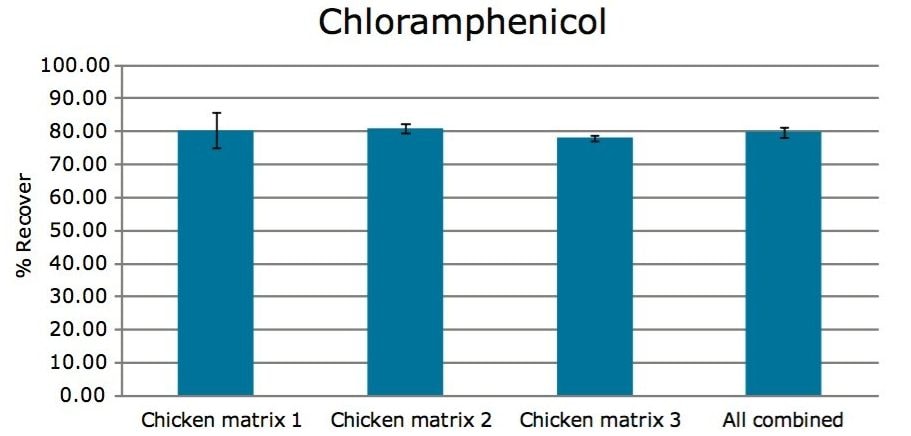
To study the recovery of chloramphenicol in chicken, three chicken breasts were purchased from different stores. Each chicken breast sample was fortified with chloramphenicol at 0.3 µg/kg, and the internal standard was 0.5 µg/kg. Fortified and blank chicken breasts were treated following the previously described sample preparation protocol. Quantitative analysis was performed with ACQUITY UPLC coupled with the Xevo TQD. The data were processed with TargetLynx Application Manager, and recoveries were calculated against the response of the non-extracted analyte. As shown in Figure 7, the average percentage recovery of chloramphenicol from three different chicken matrices was 80%.
Development of analytical methods for the detection of contaminants in food is often challenging due to the complexity of the matrix. In LC-MS/MS, co-eluting matrix components can compete with the analyte of interest during the ionization process, which can lead to ion suppression or enhancement of the analyte signal. It is therefore necessary to characterize these potential matrix effects during method development and eliminate or minimize their impact on the quantification of the analyte. Reducing matrix interference also helps to ensure method robustness. The ability to monitor matrix interferences by observing full scan background data during quantitative MS/MS experiments (RADAR) represents an important advancement in instrument design.
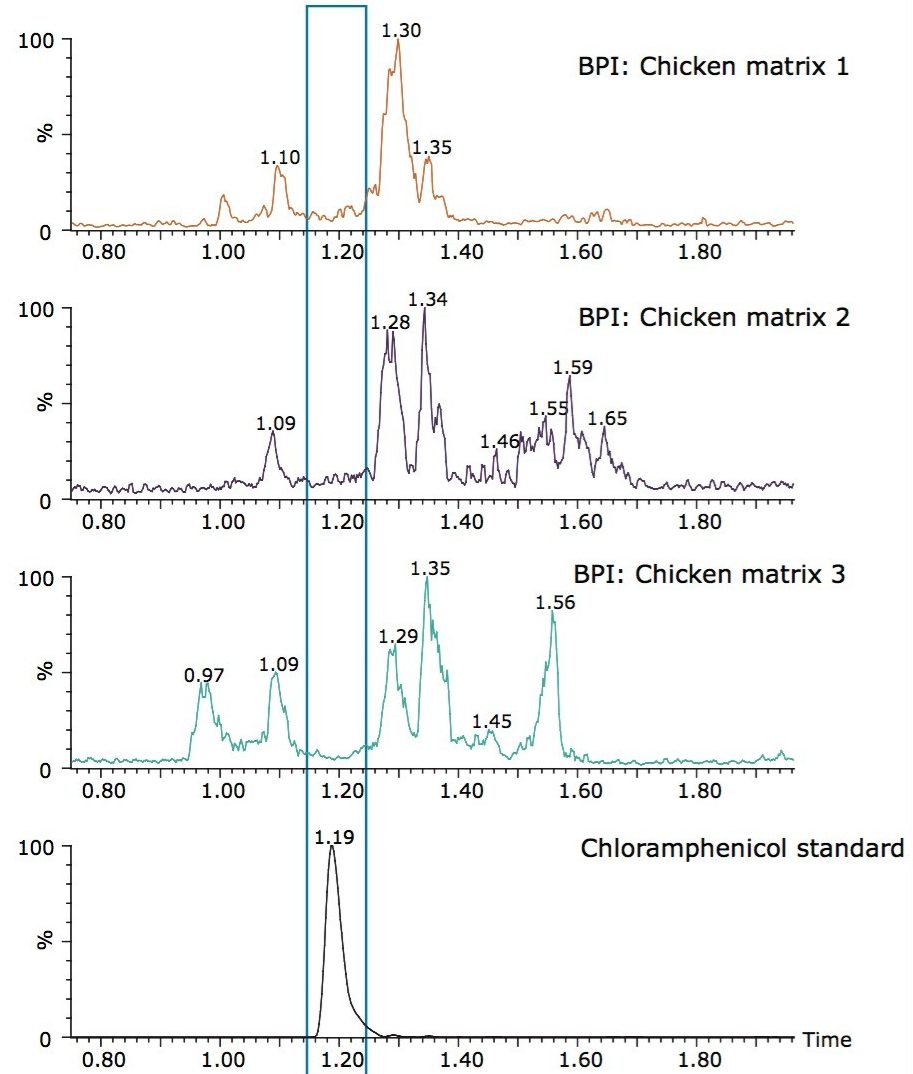
For the work presented in this application note, the Xevo TQD was operated in RADAR mode. This allowed for the simultaneous acquisition of MRMs and full scan data without any compromise in the MRM data quality or accuracy. The peak of chloramphenicol and its internal standard in the quantitative MRM chromatograms each have greater than 15 data points across the peak while simultaneously acquiring full scan data. This acquisition mode helps with making informed decisions during the process of method development and in routine analysis. Figure 8 shows the MRM chromatogram of chloramphenicol along with the full scan MS base peak ion (BPI) chromatogram for three different chicken matrices. The differences in matrix interferences among the three chicken breasts can clearly be seen. It is also apparent from Figure 8 that chloramphenicol elutes in the region that has less potential matrix interference in all three chicken samples, which leads to greater confidence in the robustness of the method. The ability to observe changes in the full scan data helps to troubleshoot any problem encountered within quantitative analyses of new samples.
This application note describes quantitative analysis of chloramphenicol in chicken breast.
720004262, March 2012