For research use only. Not for use in diagnostic procedures.
In this application note, we describe a multi-omic approach to reveal new molecular factors involved in pathogenesis of INS with potential diagnostic and therapeutic significance.
A label-free multi-omics approach for the analysis of the urine of Idiopathic Nephrotic Syndrome (INS) patients that provides both qualitative and quantitative information in a single experiment. This enables possible diagnostic and therapeutic solutions for patients with INS.
Idiopathic Nephrotic Syndrome (INS) results from the malfunction of the glomerular filter and it is the most prevalent glomerular disease in children. In spite of some progress, its pathogenesis is still unknown and therapy options are confined to gross immune modulation. A variety of methods for diagnostic and treatment purposes are available for patients; however, the lack of understanding regarding the pathogenic mechanisms underlying INS can lead to poor therapeutic response and adverse side-effects. In this application note, we describe a multi-omic approach to reveal new molecular factors involved in pathogenesis of INS with potential diagnostic and therapeutic significance.

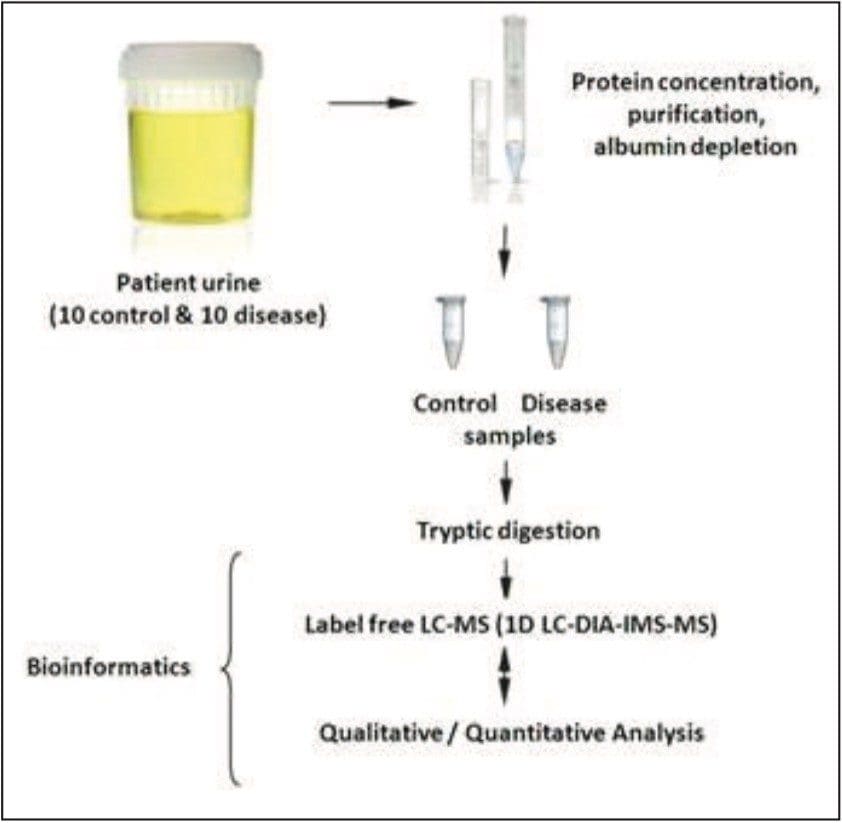
Pediatric urine samples intended for peptide analysis were prepared for LC-MS analysis as previously described.1 Samples were treated with 1% RapiGest SF prior to reduction and alkylation. Aliquots were incubated with anti-HSA resin and centrifuged using Vivaspin 5,000 MWCO filters. A series of washes using water were implemented to ensure adequate recovery. The resulting supernatant was digested using trypsin overnight as shown in Figure 2. Metabolite analysis samples were purified using Oasis HLB Extraction Cartridges. Water/methanol (90/10) washes were performed, followed by analyte elution using methanol. The resulting residue was reconstituted in 200 μL mobile phase and vortexed prior to LC-MS.
Label-free LC-MS was used for qualitative and quantitative peptide analyses. Experiments were conducted using a 90 min gradient from 5% to 40% acetonitrile (0.1% formic acid) at 300 nL/min using a nanoACQUITY UPLC System and a BEH 1.7 μm C18 reversed phase 75 μm x 20 cm nanoscale LC Column.
For metabolite identification, the LC-MS experiments consisted of a 10 min gradient from 0% to 50% acetonitrile (0.1% formic acid) at 500 μL/min using an ACQUITY UPLC System. Here, a BEH 1.7 μm C18 reversed phase 2.1 x 10 cm LC column was used.
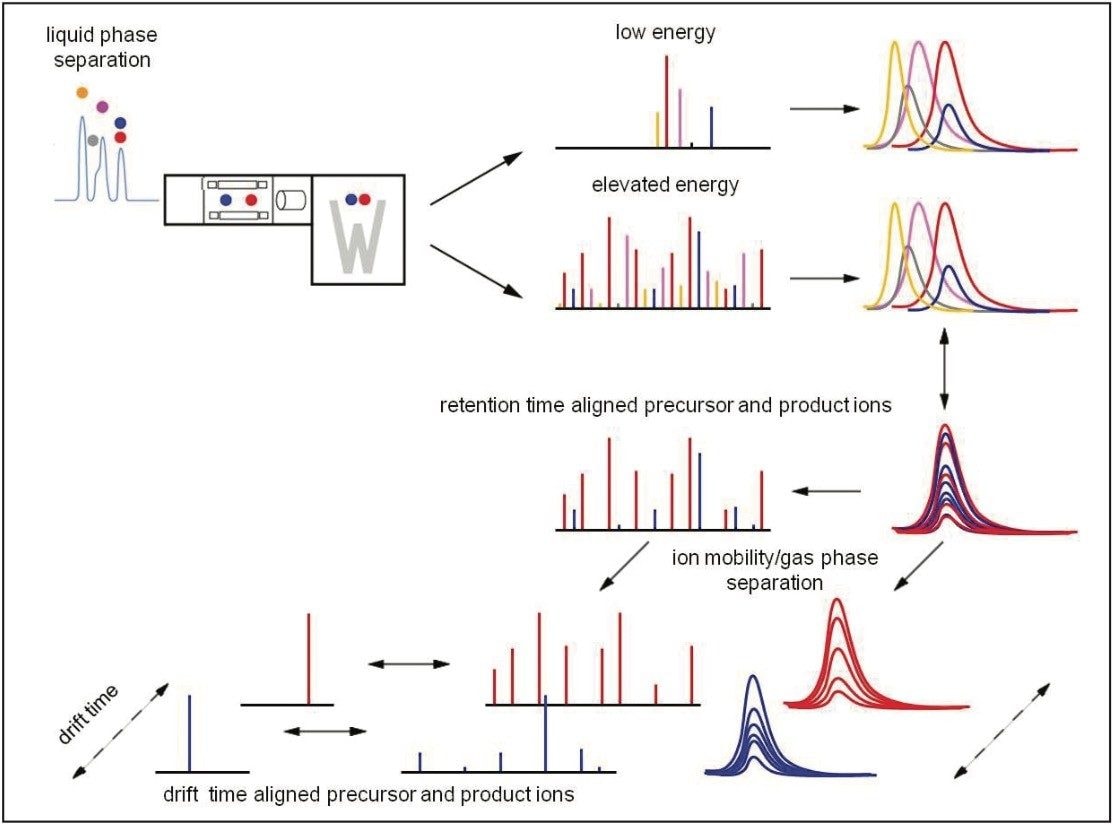
Data were acquired in data independent analysis (DIA) mode that utilized a nanoACQUITY UPLC or ACQUITY UPLC system directly interfaced to a hybrid IM-oaTof SYNAPT G2 Mass Spectrometer. Ion mobility (IM) was used in conjunction with both acquisition schemes, as illustrated in Figure 3.
The LC-MS peptide data were processed and searched with ProteinLynx GlobalSERVER. Normalized label-free quantification was achieved using TransOmics Software. The resulting metabolomic data were processed using MassLynx Software, and additional statistical analysis conducted with EZ Info.
Small amounts of the purified urine were analyzed to identify, quantify, and investigate the proteomic and metabolomic variance between control and disease pre-treated subjects.
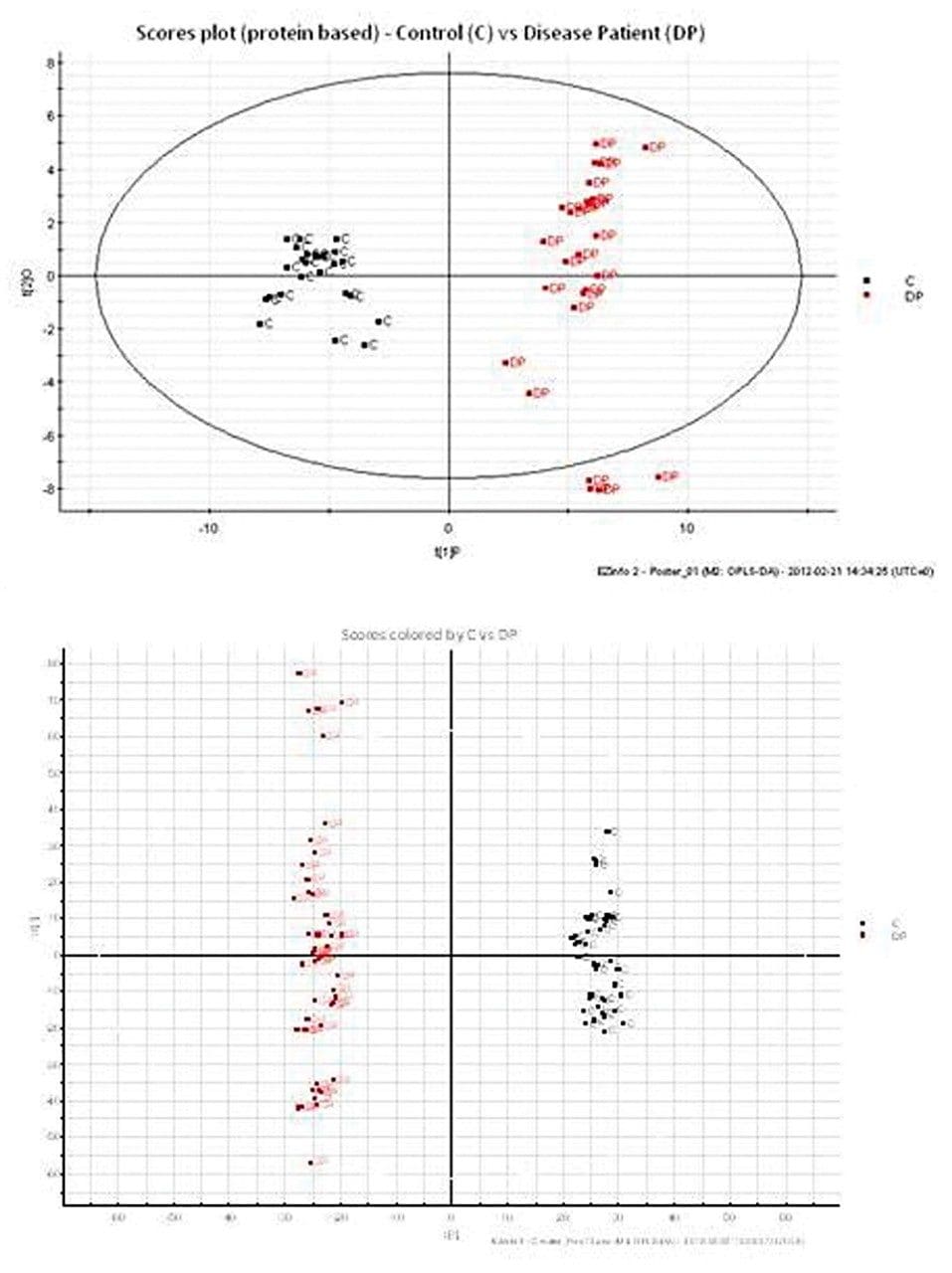
Principal Component Analysis (PCA) was used to identify and highlight significant changes between the control and disease pre-treated samples, as shown in Figure 4. Similar clustering patterns were observed for both the protein and metabolite datasets.
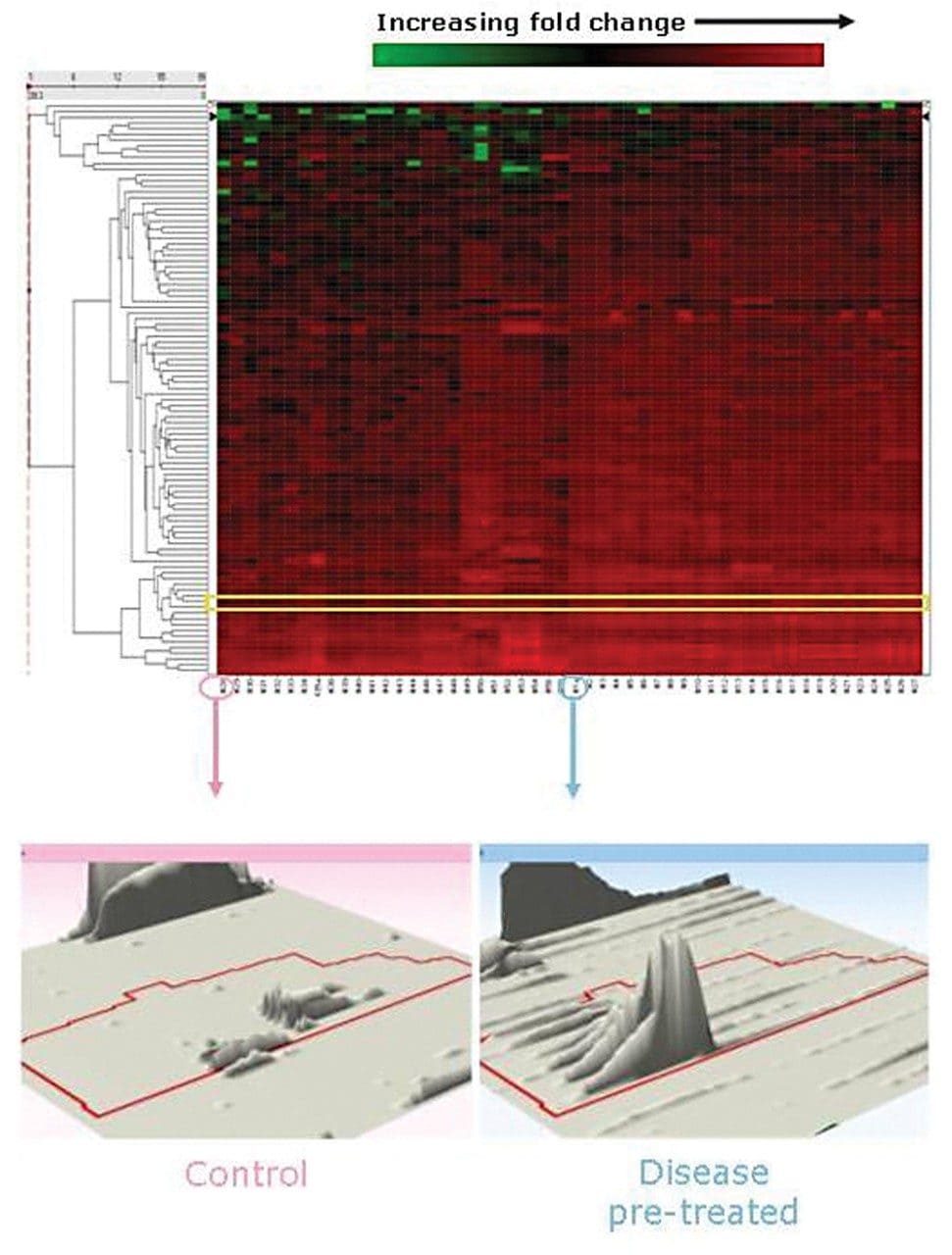
Proteomic data were aligned, normalized, and quantified. A large proportion of the identified proteins are glycosylated and over 80% of the total number of proteins identified and exhibited a significant expression fold change. Figure 5 highlights the proteins, which have greater than a two-fold change between sample sets. The fold change at the peptide level can be visually displayed using 3D montage images. A charge state feature of one of the peptides of interest is shown in Figure 5.
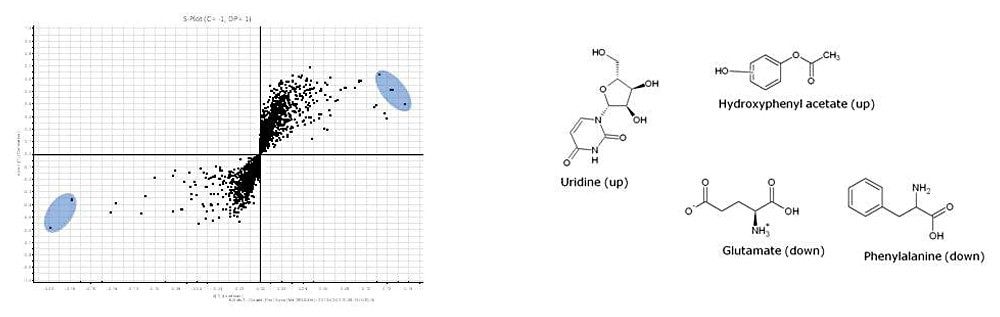
The metabolomics workflow results are summarized in Figure 6. Using the metabolite contrasting loadings plot, significant metabolite identifications can be found at the extremes and are shaded in blue. Example compounds, which were found to contribute most significantly to the variance, are also shown.
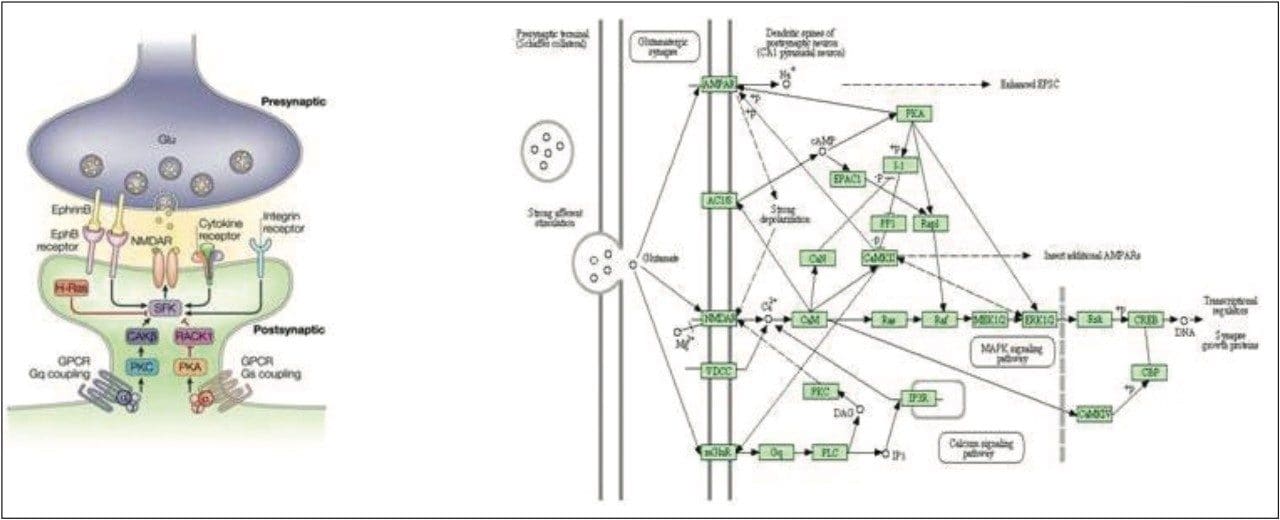
A common pathway is shown in Figure 7, which illustrates a glutamate [NMDA] receptor subunit as one such example. NMDA belongs to the glutamate-gated ion channel family of proteins and is used in neuronal system pathways. Glutamate can also be located within the same pathway. Postsynaptic Ca(2+) is thought to increase through the NMDA receptors, which activate several signal transduction pathways including Erk/MAP kinase and cAMP regulatory pathways.
720004329, May 2012