This Application note demonstates UPLC/FLR analysis under the control of an Empower workstation, and used with the GlyoBase 3+ database for glycan unit assignment, represents a powerful analytical platform for analyzing the oligosaccharides attached to glycoproteins.
ACQUITY UPLC/FLR analysis under the control of an Empower workstation represents a powerful and fully integrated analytical platform for analyzing the oligosaccharides attached to glycoproteins.
Glycosylation is the most complex post-translational protein modification. More than half of all proteins are glycosylated, and, to elucidate their function, it is important to define the structures of the glycans that are covalently attached to their surfaces. The manufacturing of glycoprotein life sciences products can be challenging and a number of factors can have a major impact upon their glycosylation, including the cell type in which the protein is expressed, media composition, and processing parameters such as dissolved oxygen, pH, the carbon source, and temperature.
The glycosylation of biotherapeutics is often a critical product attribute for therapeutic efficacy and safety. Fluctuations in the process can put product integrity at risk and therefore it is important to monitor glycosylation accurately throughout the process.
In collaboration with Waters, the National Institute for Bioprocessing Research and Training (NIBRT), based in Ireland, has developed a glycan database for use in conjunction with UPLC glycan separations.

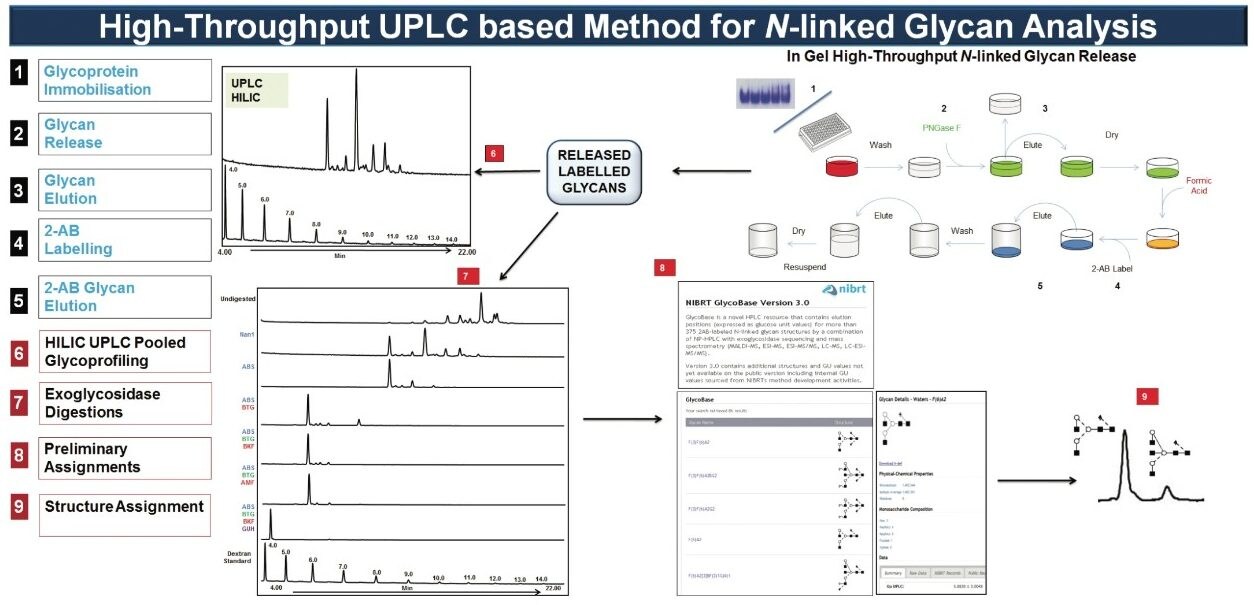
The analytical platform utilized by NIBRT is described in Figure 1. Briefly: N-glycans are released by PNGase F and fluorescently labeled with 2-aminobenzamide before separating the labeled glycan pool by hydrophilic interaction chromatography (HILIC).
The Waters ACQUITY UPLC System applies an optimized hydrophilic interaction chromatographic separation (HILIC) using columns containing sub-2-μm particles. The combination of the ideal selectivity of diol-bonded Bridged Ethyl Hybrid (BEH) particles and exceptional peak capacity at higher flow rates results in overall increases in speed, sensitivity, and resolution compared to standard HPLC systems.
The monoclonal antibody Herceptin was immobilized in acrylamide gel blocks and N-linked glycans released using peptide-N-glycosidase F. The glycan pool was labelled with 2-aminobenzamide (2AB). A linear gradient of 50 mM ammonium formate buffer, pH 4.4, and acetonitrile was used for glycan separation.
|
UPLC system: |
ACQUITY UPLC |
|
Column: |
ACQUITY UPLC BEH Glycan Column (2.1 x 150 mm) |
|
Column temp.: |
40 °C |
|
Sample temp.: |
5 ± 5 °C |
|
Flow rate: |
0.561 mL/min |
|
Mobile phase A: |
50 mM ammonium formate (pH 4.4) |
|
Mobile phase B: |
100% acetonitrile |
|
Time (min) |
Flow rate |
%A |
%B |
Curve |
|---|---|---|---|---|
|
1. Initial |
0.561 |
30.0 |
70.0 |
6 |
|
2. 1.47 |
0.561 |
30.0 |
70.0 |
6 |
|
3. 24.81 |
0.561 |
47.0 |
53.0 |
6 |
|
4. 25.50 |
0.400 |
70.0 |
30.0 |
6 |
|
5. 26.25 |
0.400 |
70.0 |
30.0 |
6 |
|
6. 26.55 |
0.400 |
30.0 |
70.0 |
6 |
|
7. 28.55 |
0.561 |
30.0 |
70.0 |
6 |
|
8. 32.00 |
0.400 |
30.0 |
70.0 |
6 |
|
UPLC detector: |
ACQUITY UPLC FLR Detector |
|
Wavelengths: |
λex =330 nm, λem = 420 nm |
|
Data rate: |
10 pts/sec |
|
PMT gain: |
20 |
|
Time constant: |
Normal |
|
Auto Zero On Injection Start (check) |
Empower Software
GlycoBase 3.0 database
The preliminary assignment of glycan structures is obtained by interrogating the NIBRT database(http://glycobase.nibrt.ie) in which peak retention times are expressed in glucose units (GU values) by alignment to a dextran hydrolysate ladder. Chromatographic resolution and reproducibility play a vital role in obtaining accurate and reproducible GU measurements and quantitative data. In the ACQUITY UPLC System, the GU value standard deviation is below ± 0.01 GU from 8 to 10 independent analyses, illustrating the superb peak resolution and run-to-run reproducibility.
An ACQUITY UPLC System combined with a BEH glycan column provides significant enhancement in terms of peak capacity and resolution in comparison to classical HPLC technologies.
In Figure 2, a 30-minute separation of N-glycans from human serum on a (1.7-µm) BEH glycan column is compared with chromatograms that were generated using a classical HPLC system with amide columns (5-µm and 3-µm particle size).
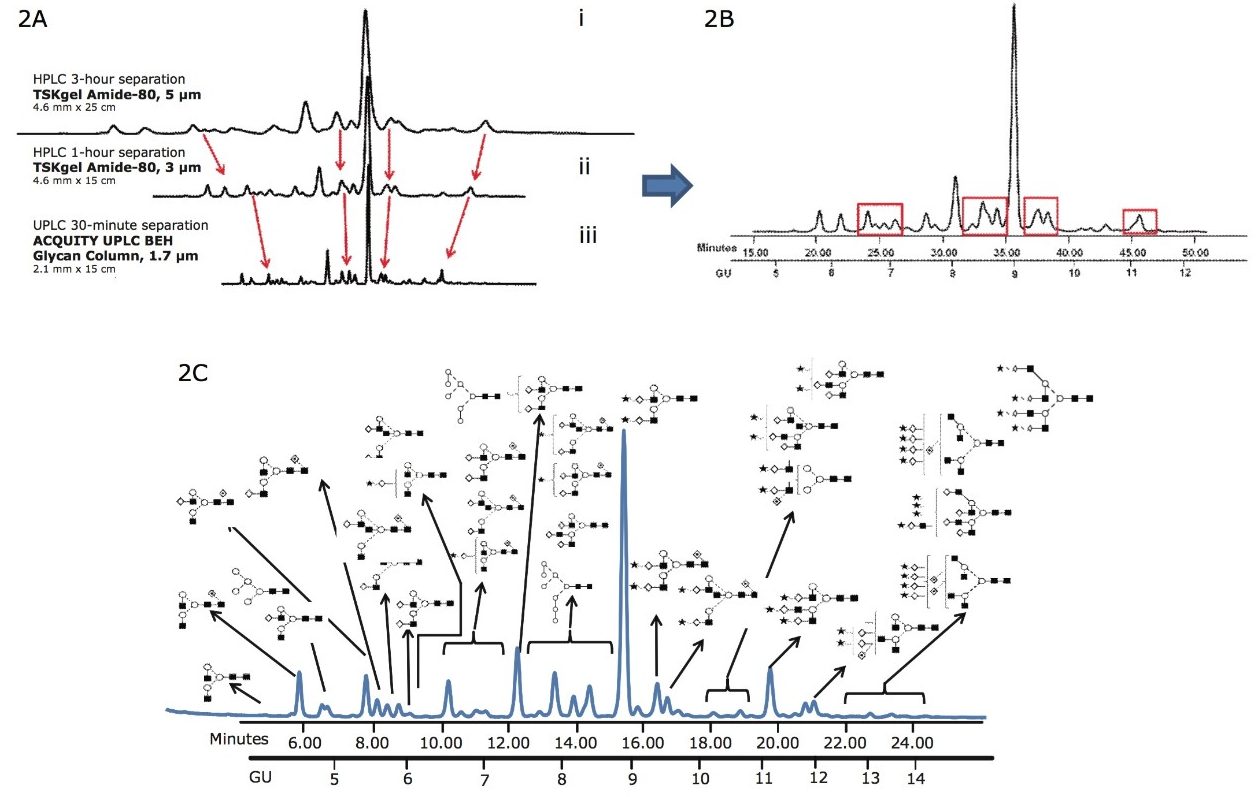
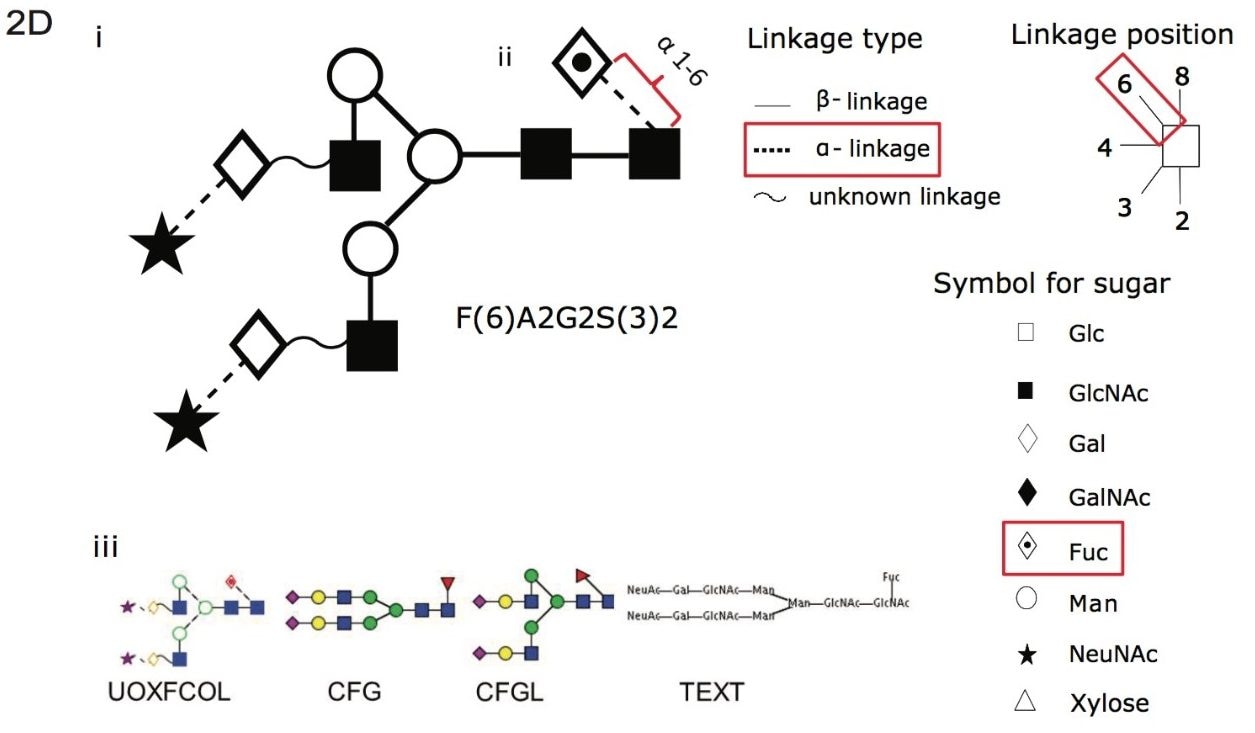
Figure 2A. Comparison of the 3-hour HPLC method and 1-hour HPLC separation of complex human serum glycans on (i) 5-μm and (ii) 3-μm TSK-GEL Amide-80 columns using a 30 minute Waters UPLC method (iii) BEH glycan column 1.7-μm, 2.1 mm x 15 cm.
Figure 2B. Separation of human serum protein glycans with classical HPLC methods, TSK-GEL 3-μm Amide-80 column, 1 hour run time.
Figure 2C. The same human serum glycome analyzed on Waters ACQUITY UPLC. The most abundant of the 136 glycans that were identified are annotated (Figure 2D) There is significant decrease in run times and improved separation of the glycans compared with the classical HPLC methods.
Figure 2D. (i) A cartoon depicting the linkage type, linkage position and symbol for common sugars used with the Oxford notation. (ii) An example using F(6)A2G2S(3)2, where core fucose F(6) α 1-6 linkage is depicted with Oxford notation highlighted in red for linkage type, linkage position and symbol for associated sugar. (iii) Glycobase has a conversion function (developed by Eurocarb DB) that can represent glycans in other common formats as can be see with F(6)A2G2S(3)2.1,2
Figure 3 shows that the ACQUITY UPLC System rapidly resolves glycans that were released from Herceptin in 12 minutes. 24 glycans were identified and quantified using the NIBRT database and exoglycosidase array digestions; the major glycan species are annotated in the figure.
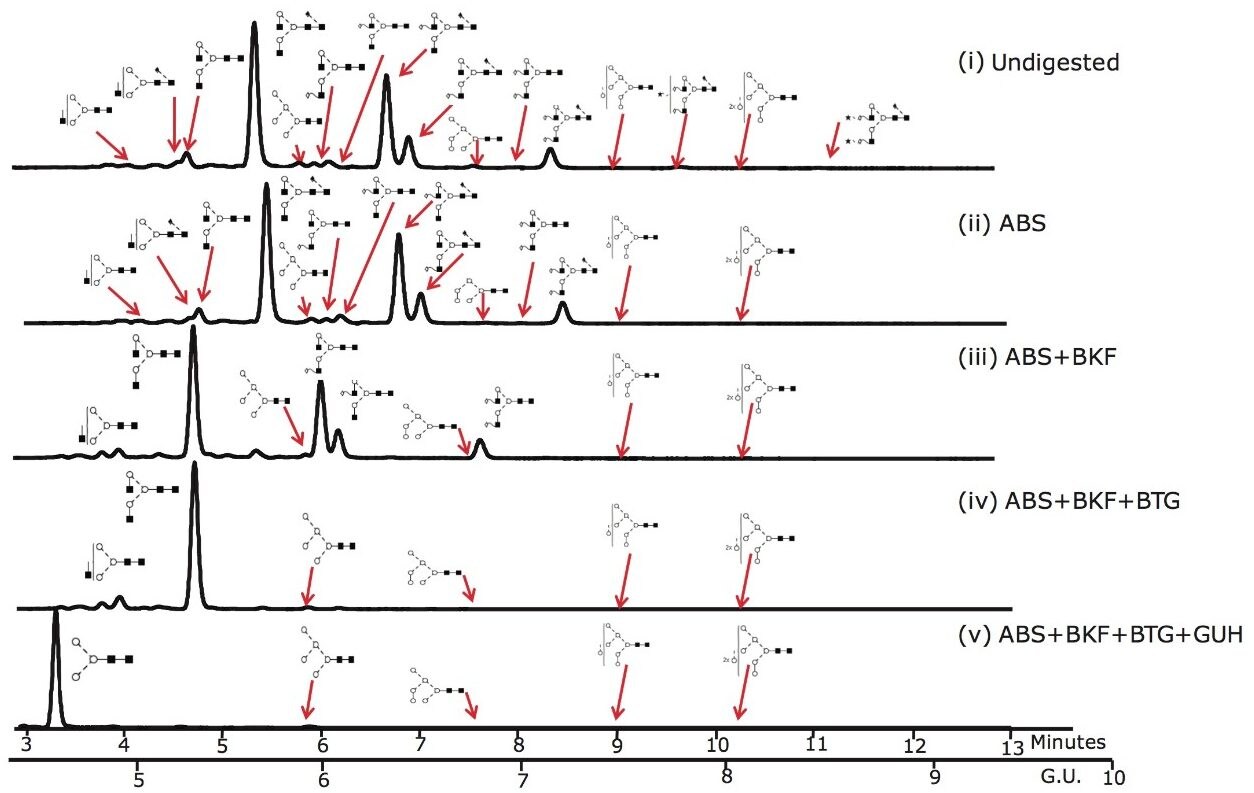
Figure 3. UHPLC analysis of the pool of glycans released from Herceptin (Trastuzumab) monoclonal antibody and the exoglycosidase array digestions that provide monosaccharide sequence and linkage information. 10 ug of Herceptin was used for the initial release by PNGaseF and intact pool represents approximately 2% of the total released N-glycan pool.
(i) Intact N-glycan pool released by PNGase F; (ii) ABS, Arthrobacter ureafaciens alpha-sialidase (1 U/mL) digestion. ABS releases α2-3 /6/8 sialic acids; (iii) ABS + BKF, bovine kidney alpha-fucosidase (BFK) (1 U/mL) array digestion. BKF releases α1-6/2 fucose (iv) ABS +BKF+ BTG, bovine testes beta-galactosidase (BTG) (1 U/mL) array digestion. BTG releases galactose β1-3>1-4 linkages and (v) ABS+BKF+BTG+N-acetylglucosaminidase (GUH) (4 U/mL) array digestion.
Included in GlycoBase 3.0 (http://glycobase.nibrt.ie) is the Waters collection of UPLC GU values of the glycans of a diverse group of nine samples (Herceptin, Human and Mouse serum IgG, recombinant Erythropoietin, Bovine Fetuin, Bovine Ribonuclease B, Yeast Invertase and Human serum).These values were obtained by the systematic analysis of released glycans using Waters HPLC and UPLC technologies and the NIBRT glycan analytical platform. The profiles of the glycan pools and the enzymatic digestions can be obtained from a link (http://glycobase.nibrt. ie/glycobase/documents/enzyme.pdf). In brief, the N-linked glycan pool is separated equally into 5 (0.5 mL) eppendorfs and appropriate exoglycosidase enzymes are sequentially added with 10x 50 mM sodium acetate pH 5.5 buffer to build up the array. These are labeled (i) undigested control; (ii) ABS; (iii) ABS+BFK; (iv) ABS+BFK, BTG; and finally (v) ABS+BFK, BTG, GUH. Enzyme nomenclature and amounts are described below.
720004202, September 2012