Using the UNIFI API to Enable Streamlined Data Export to Protein Metrics Byos Platform for Processing of Biotherapeutic Protein and Peptide LC-MS Data
Dies ist ein Applikationsbericht, der keinen detaillierten Abschnitt zu Versuchen enthält.
Abstract
Employ the Application Programming Interface (API) in UNIFI™ Scientific Information System v1.9.4 to enable third-party applications such as the Byos® platform (Protein Metrics, Inc., Cupertino, CA, USA) to access, read, and process data acquired by UNIFI.
Introduction
Waters™ UNIFI-based LC-MS system solutions for biopharmaceutical analysis excel at automating the generation, processing, review, and reporting of biopharmaceutical characterization and monitoring data, including foundational tools for operating within compliant environments. Using the API functionality within UNIFI, analysts can further benefit from extended characterization capabilities offered by third-party software vendors. Byos, the first UNIFI API partner in the biopharmaceutical space, has created a vendor-neutral informatics platform of protein characterization tools designed to enable laboratories to streamline data comparisons across multiple vendor LC-MS platforms, and provide access to extended characterization workflows, supplementing those provided by Waters and other vendors. This API functionality allows the Byos application secure permissions-based access to the UNIFI database to extract raw data acquired by UNIFI, for use by the tools within the Byos software suite. This facilitates streamlined integration of UNIFI data with that from other data sources for commonplace workflows, such as intact/subunit protein mass and peptide mapping, as well as access to extended characterization tools for workflows, such as protein crosslinking analysis, not currently addressed in UNIFI or MassLynx™ today. Waters and Protein Metrics have developed a streamlined workflow for generating data in Byos-compatible formats, which circumvents the need for manually exporting data to generic file formats for subsequent manual import into the Byos platform, enabling a better user experience on both platforms.
Results and Discussion
The Solution
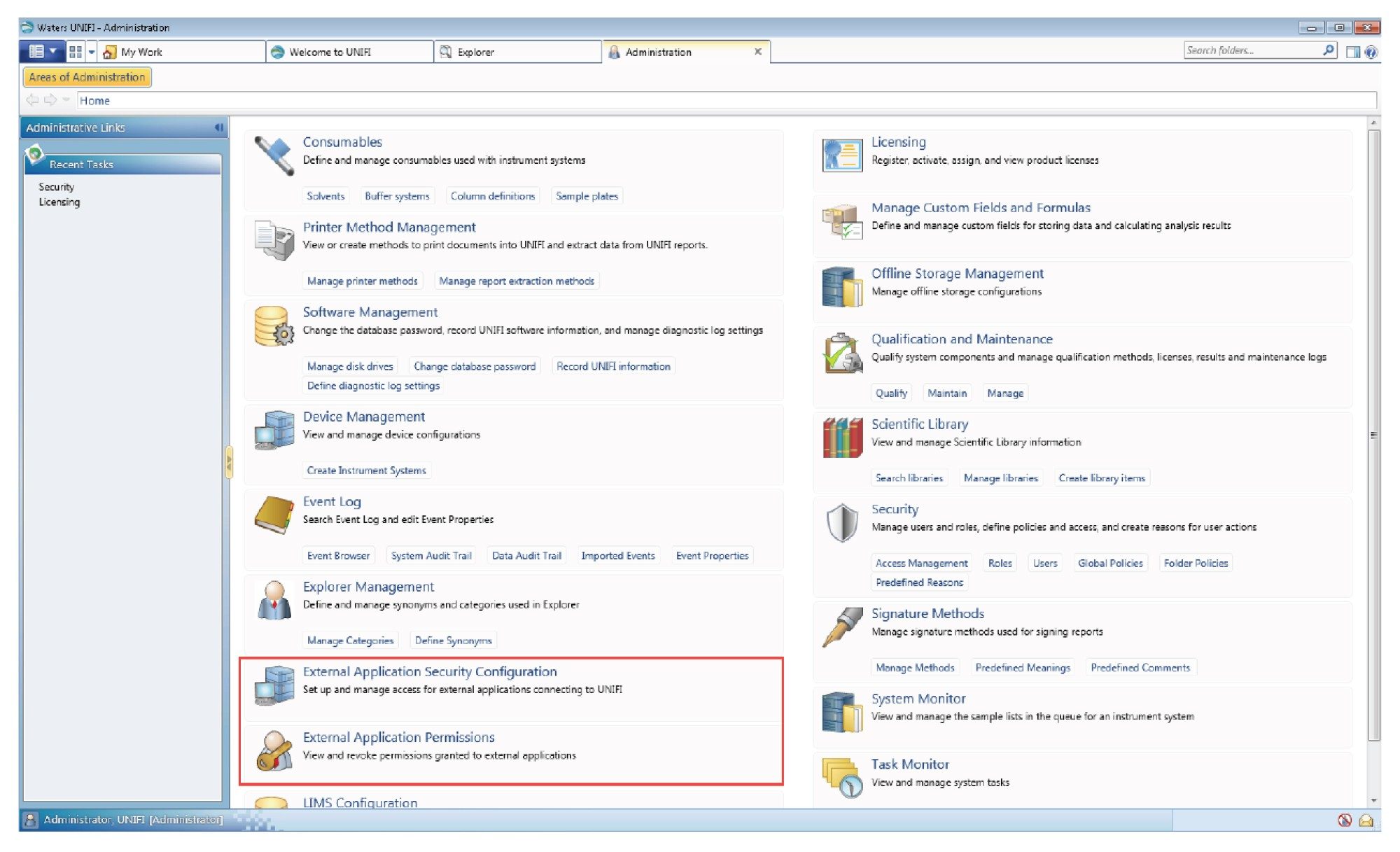
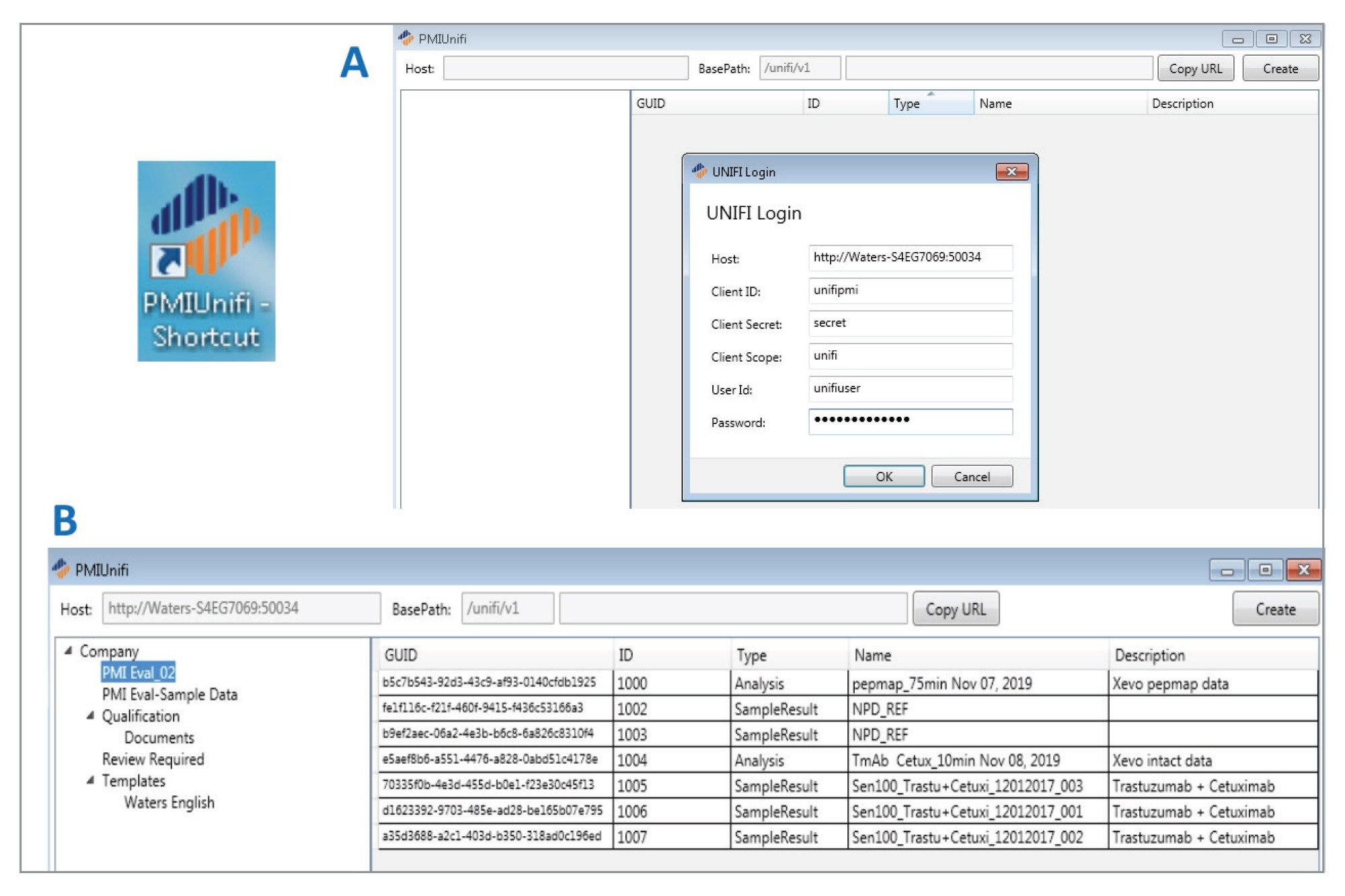
To demonstrate the combined UNIFI/Byos workflow, we have chosen to analyze intact trastuzumab and trypsin-digested NIST mAb samples from data collected on a Waters Xevo™ G2-XS Tof Mass Spectrometer under UNIFI v1.9.4 control. Figure 1 shows two additional elements on the UNIFI Administration tab (External Application Security Configuration and External Application Permissions) that facilitate third-party secure access to UNIFI acquired data. Once PMIUNIFI is added to the approved client list, with established login credentials, the UNIFI database can be accessed through this interface and Byos-compatible Byspec2 files can be generated (Figure 2).
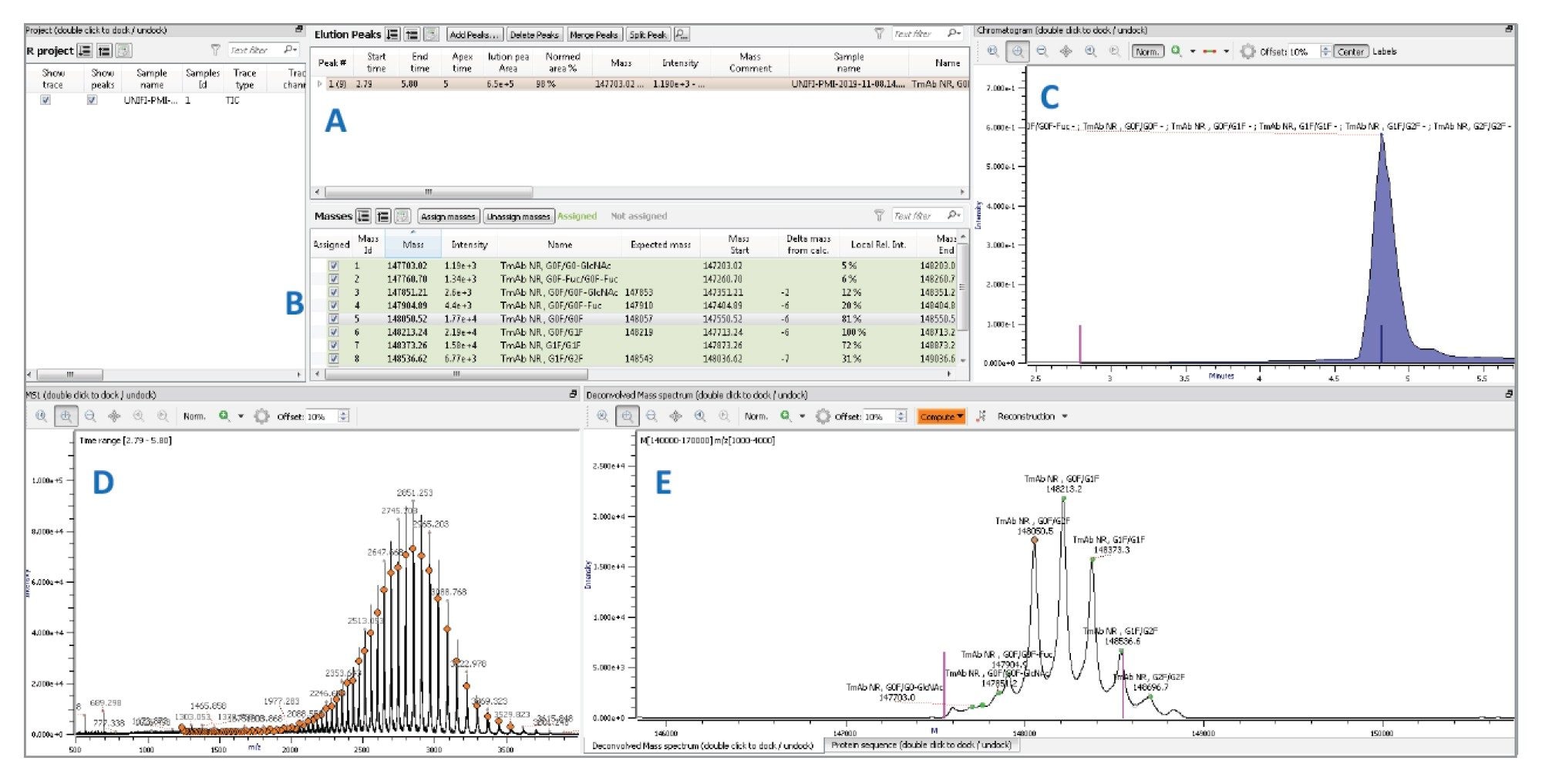
Within the Byos platform, an analysis workflow, including expected protein sequences and potential modifications, is established and used to process the selected data files and generate reports. The results for trastuzumab intact mass analysis (Figure 3) using the Byos Intact workflow were found to be comparable to those generated within UNIFI.
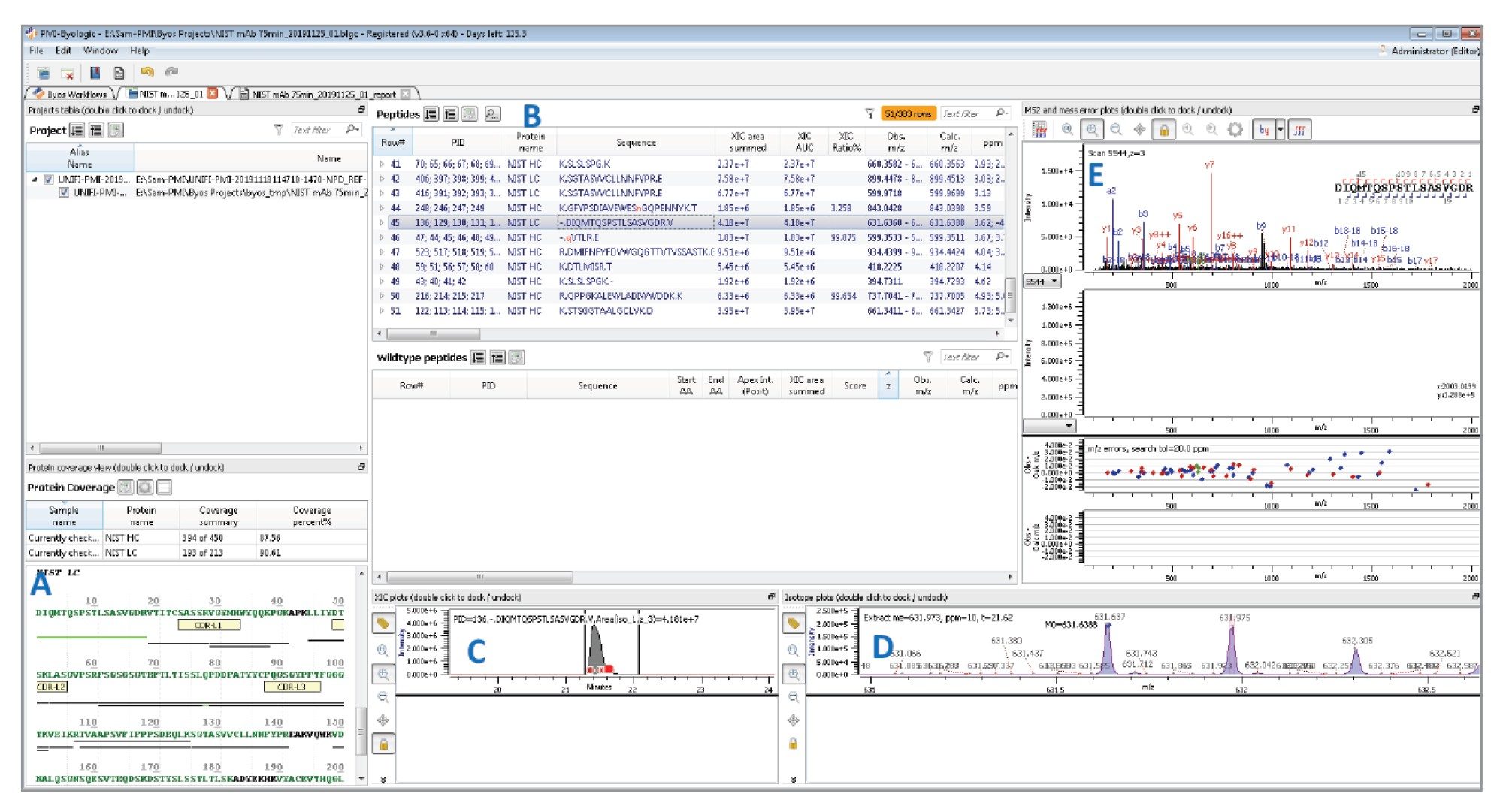
Peptide workflow results for a typical NIST mAb tryptic digest acquired by UNIFI (Figure 4) include displays demonstrating high sequence coverage (Figure 4A), and the list of matched peptides (Figure 4B) with strong MS fragmentation scores calculated from data-independent MSE fragmentation data. Again, the quality of the processed Byos data appears comparable to that obtained with the UNIFI peptide mapping workflow.
Conclusion
The UNIFI API enables third-party applications such as Byos to take advantage of high-quality Waters UPLC™-HRMS (high-resolution mass spectrometry) data generated on the UNIFI Informatics platform via streamlined and secure access to the UNIFI database. This circumvents the need to manually export files from UNIFI in order to utilize third-party software packages. Biopharmaceutical characterization labs will now be able to take maximum advantage of both UNIFI and Byos data workflows for routine and extended characterization of biomolecules.
720006743, January 2020