
This Application brief demonstrates a new Application Programming Interface (API) released in UNIFI 1.9.2 enables third party applications to read and process data acquired on instruments using the UNIFI platform. We describe the integration of Mass-MetaSite and WebMetabase with UNIFI in order to identify drug metabolites in hepatocyte incubations.
Third party applications have the potential to add significant value to the native capabilities of the UNIFI Scientific Information System. Here, we describe the integration of Mass-MetaSite and WebMetabase to process drug metabolism data in a drug discovery environment for discovery applications.
Mass-MetaSite and WebMetabase (Lead Molecular Design, Barcelona, Spain) represent a suite of applications targeted at addressing a fundamental limiting issue in drug discovery – designing out metabolic liabilities in order to bring forward compounds into development that are likely to have acceptable pharmacokinetics in humans. High resolution, ion mobilityenhanced mass spectrometry, combined with UPLC and UNIFI, excels at generating datasets, which can be used to identify drug metabolites. Mass-MetaSite and WebMetabase complement this by providing additional functionality including:
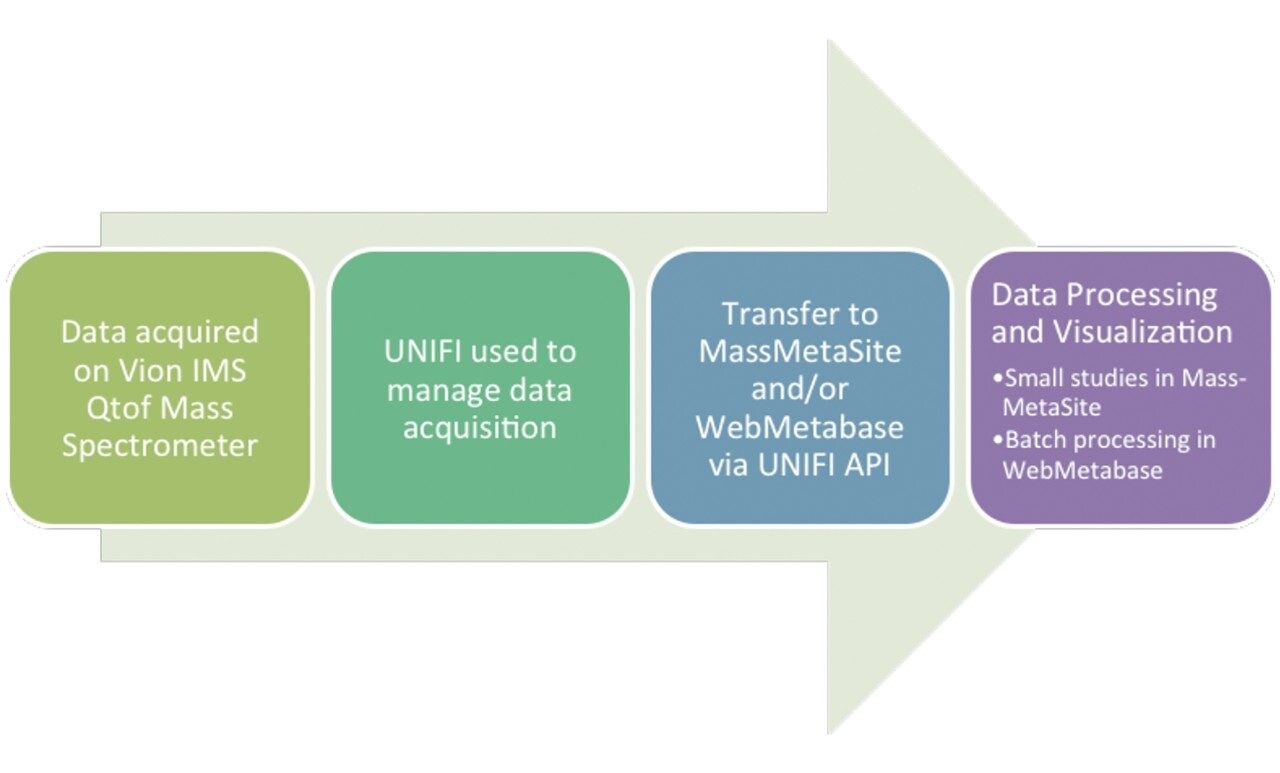
Waters and Lead Molecular Design developed a seamless workflow in which UPLC/HDMSE data are acquired in UNIFI, then transferred, processed, and visualized within Mass-MetaSite and/or WebMetabase. The combination of Vion IMS QTof Mass Spectrometer, UNIFI, and High Definition MSE acquisition schema affords a user-friendly, robust, generic, and selective approach to the characterization of complex samples.
Verapamil was incubated in a rat hepatocyte suspension at a 10 μM substrate concentration and aliquots taken at defined time points, quenched with acetonitrile (2 volumes acetonitrile:1 volume of incubation), centrifuged, and the supernatant transferred to a 96 well plate for subsequent analysis. Samples were further diluted 1 in 10 with urine. HDMSE datasets were acquired on a Vion IMS QTof Mass Spectrometer equipped with an ACQUITY UPLC I-Class System, running under UNIFI control. The general workflow for the solution is shown in Figure 1. Acquired datasets were transferred onto a processing workstation running UNIFI 1.9.4 and UNIFI API 1.0, though it is also possible for data to be processed directly from the acquisition computer. Secure access by Mass-MetaSite and WebMetabase to the UNIFI system was achieved by password-based authentication. Subsequent selection of samples for downstream processing within Mass-MetaSite was achieved by navigation within the UNIFI folder structure (Figure 2).
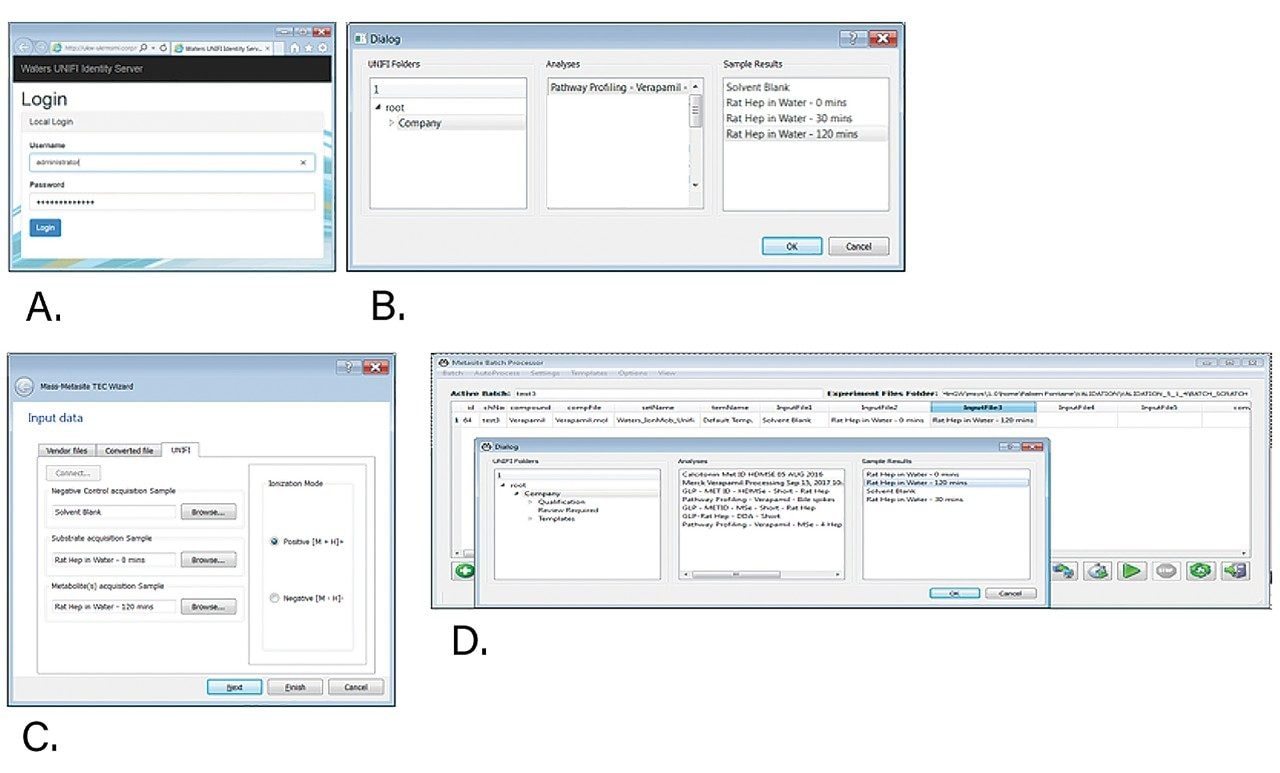
Two modes of data processing are possible, either single sample processing with comparison to a defined control within Mass-MetaSite or batch processing of multiple compounds with multiple conditions (species, time, matrix) within WebMetabase (Figure 2). Data review can be achieved in a standalone mode within the application, or as a cloud-enabled solution within a standard web browser.
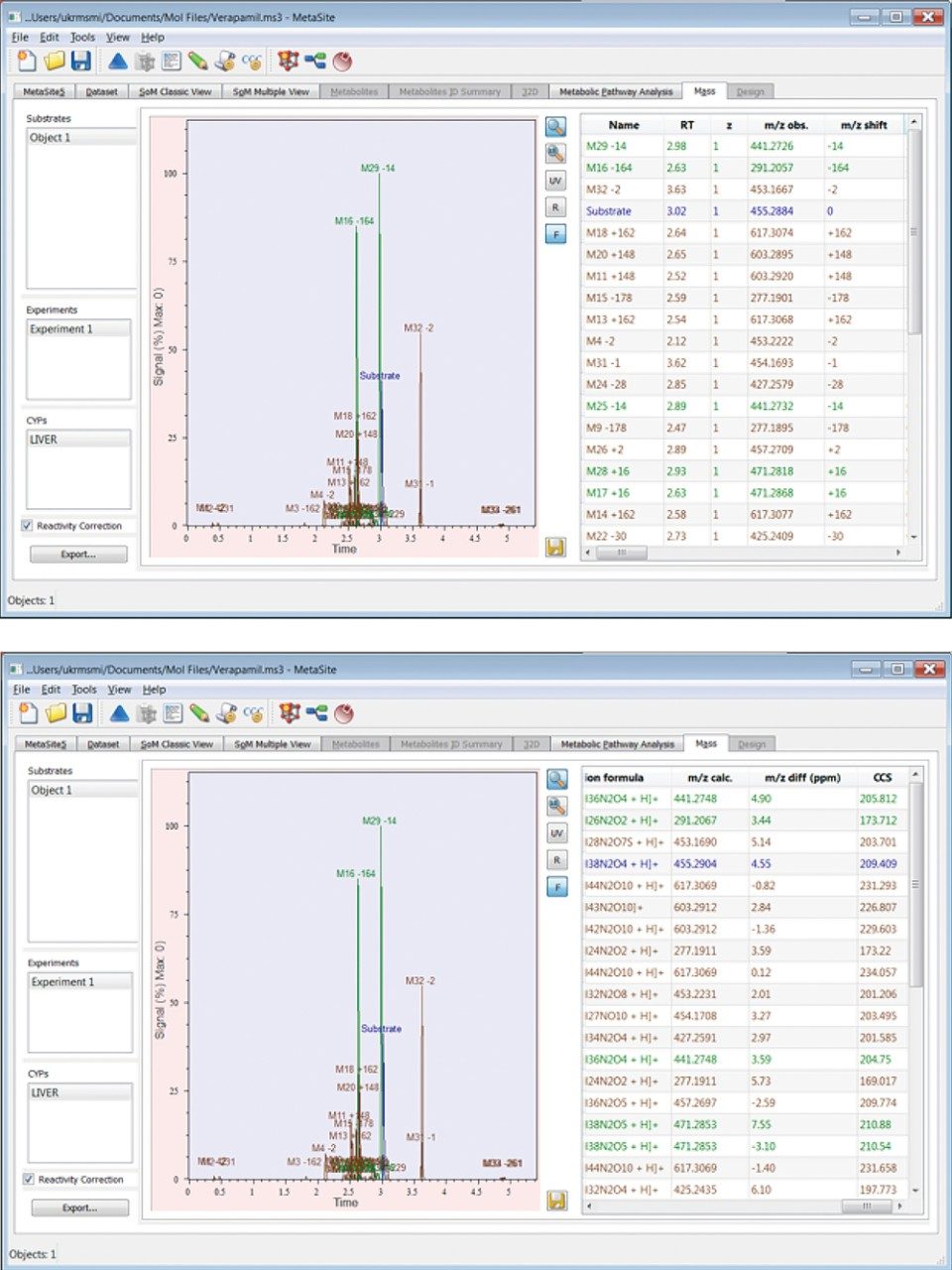
Representative outcomes within Mass-MetaSite for the 30 minute time point of the verapamil incubation are shown in Figure 3. Within UNIFI, metabolites are characterized not only on the basis of m/z and retention time, but with their collision cross section (CCS), allowing isobaric metabolites to be distinguished across experiments. Like UNIFI, Mass-MetaSite generates product ion spectra for identified metabolites by utilizing drift time correlation, resulting in clean spectra comparable in quality to those obtained with targeted MS/MS. A formal validation of the outcomes from data processed within both Mass-MetaSite and UNIFI confirmed that m/z and CCS values were consistent.
For CCS values, there was an average difference between UNIFI and Mass-MetaSite of 0.04%. For m/z values, there was an average difference between UNIFI and Mass-MetaSite of 0.05 ppm.
The UNIFI API provides a platform by which third party applications can take advantage of the exceptional data quality afforded by UPLC-HR-IMS-MS, enabling a rich ecosystem of software to thrive. The capabilities of Mass- MetaSite and WebMetabase are complementary to those offered by UNIFI, and afford access to a comprehensive toolset which can be used to understand the metabolism and optimize the pharmacokinetics of lead compounds within a drug discovery environment.
CCS (Collision Cross Section): Value describing an ion’s structure, charge, and conformation, determined by the interaction of an individual ion with a neutral gas in an ion mobility drift cell. CCS is directly related to drift time.
IMS (Ion Mobility Spectrometry): A separation technique in which ions are separated based on their size, shape, and/or charge as they pass through a neutral gas. IMS is frequently coupled with mass spectrometry as IMS-MS.
HDMSE (High Definition MSE): Data-independent acquisition method combining ion mobility separation and MSE data acquisition. Low collision energy IMS-MS scans and high collision energy IMS-MS scans are alternated, allowing both precursor and product ion accurate mass measurements to be obtained.
720006362, July 2018