For research use only. Not for use in diagnostic procedures.
This is an Application Brief and does not contain a detailed Experimental section.
Introduction of the desorption electrospray ionization (DESI) technique coupled to mass spectrometry imaging (MSI) using a Xevo G2-XS Quadrupole Time-of-flight (QTof) Mass Spectrometer for distinguishing tissue types in breast samples for clinical research.
Over the past decade, mass spectrometry imaging has been used increasingly by researchers to investigate the distribution of metabolites, drugs, peptides, and proteins at tissue surfaces. The potential for the application of MSI to unambiguously map hundreds of biomolecules in a single analysis has led to this approach being used in research studies of cancer. Recently, there has been a significant increase in the application of desorption electrospray ionization (DESI) because this soft ionization technique can be performed under ambient environmental conditions. Furthermore, it requires little to no sample preparation and is minimally invasive, making it suitable for direct tissue analysis. DESI-MSI has potential to provide non-subjective information about biochemical distribution of molecules after just one measurement. Therefore, this technique enables robust tissue recognition and identification of tissue-specific lipid ion patterns, which could, in the future, be useful in cancer diagnosis and prognosis at a histology-level. DESI-MSI is compatible with both the Waters SYNAPT G2-Si and Xevo G2-XS Mass Spectrometers.
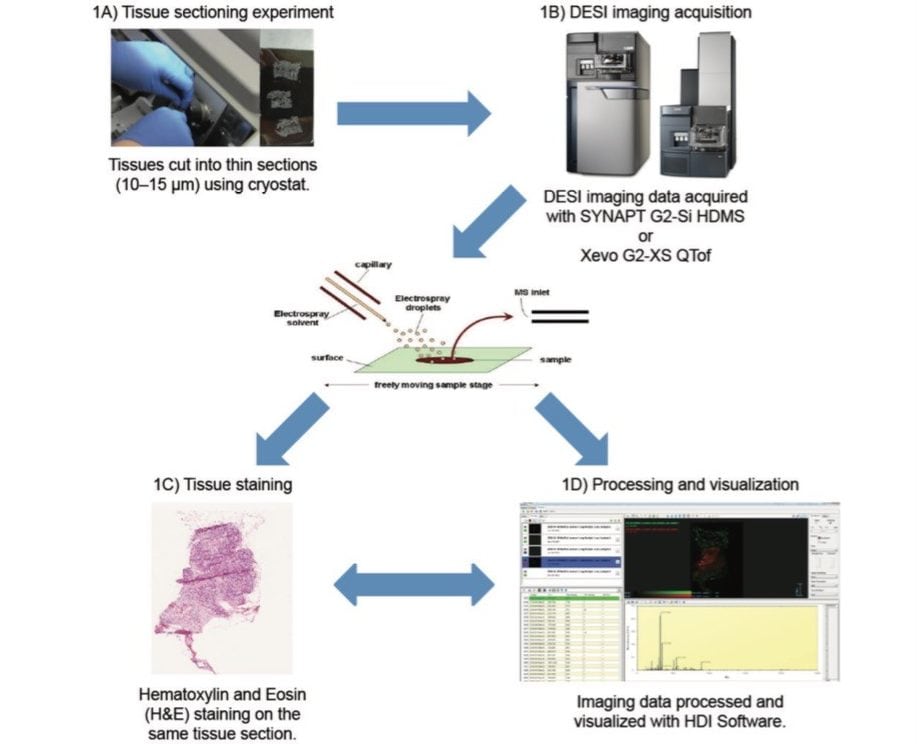
In order to perform a DESI-MSI experiment on breast tissue, a fresh frozen 10 µm tissue section was mounted onto a glass slide (Figure 1A). The slide was placed directly onto the linear moving stage of the Prosolia 2D DESI source without any other pre-treatment. An optical image was recorded and co-registered within the High Definition Imaging (HDI) Software, version 1.4 (Figure 1D). This optical image was then used to define the area to be imaged. A typical DESI-MSI experiment involves rastering the tissue sections with an ionization beam, line by line, to acquire mass spectra at defined x and y coordinates. Each sampling location corresponds to a unique mass spectrum. For DESI-MSI of the breast samples, pixel size in the x-direction is defined at 100 µm and the acquisition rate was set to five scans per second using the solvent mixture of 95:5 methanol:water.
Following DESI-MSI analysis, the tissue sections were Hematoxylin- and Eosin- (H&E) stained (Figure 1C) made possible because DESI is a non-destructive technique. Overlaying the H&E-stained optical image with the DESI-MS ion images provides a topographical comparison of the biochemical features of the same section.
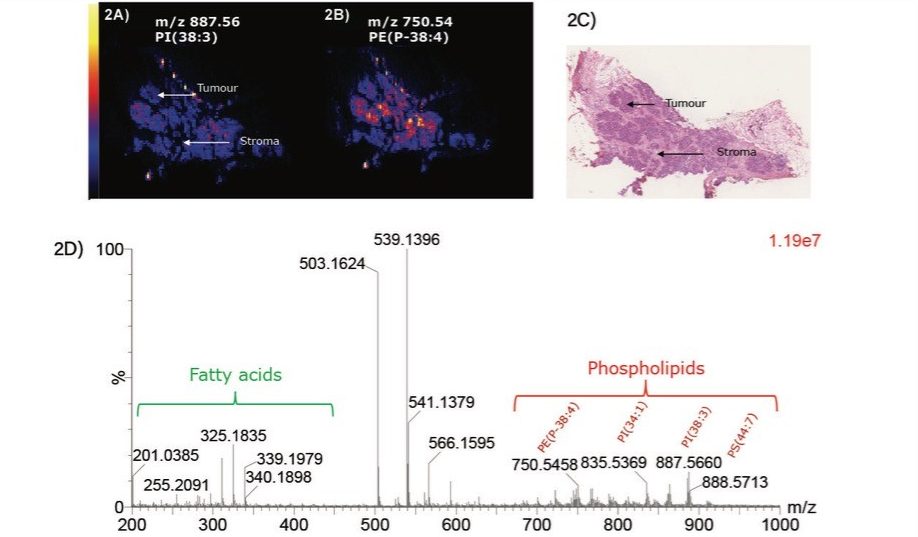
In terms of breast tissue analysis, DESI has shown a very good separation of the malignant component from the rest of the tissue section as shown in Figure 2 where PI (38:3) corresponding to m/z 887.56 is characteristic of the malignant part of the tissue. DESI generates intense fatty acid and phospholipid signals in negative ionization mode as shown in Figure 2.
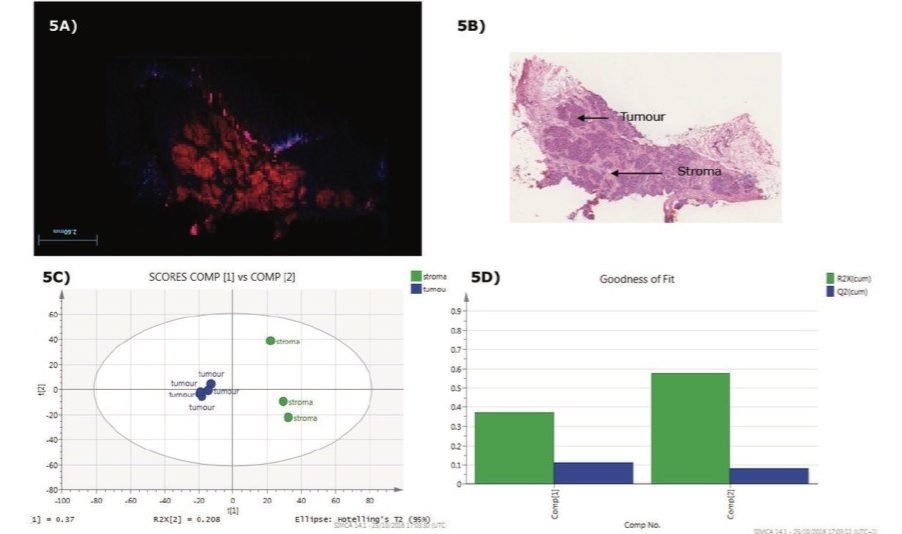
Statistical analysis was performed from HDI processed DESI-MSI data by first defining the Regions of Interest (ROIs) on the MSI ion images based on histopathological annotations. Differences between the two tissue types found (stroma and tumor) were evaluated. Figure 3 shows principal component analysis (PCA) using SIMCA software (Umetrics), which demonstrates a good fit for the model generated with a high R2 (Regression fitness) and Q2 (Predictive fitness). Using the SIMCA informatics workflow, it is possible to link the ion images of the dataset with the loading plot in HDI.
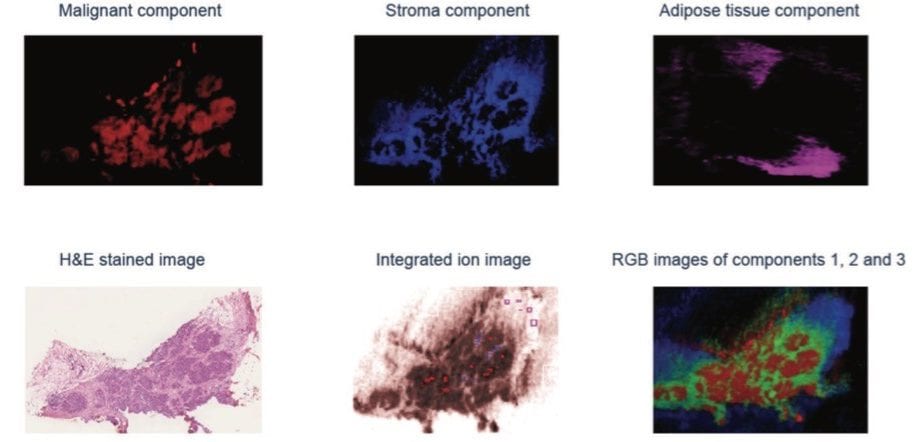
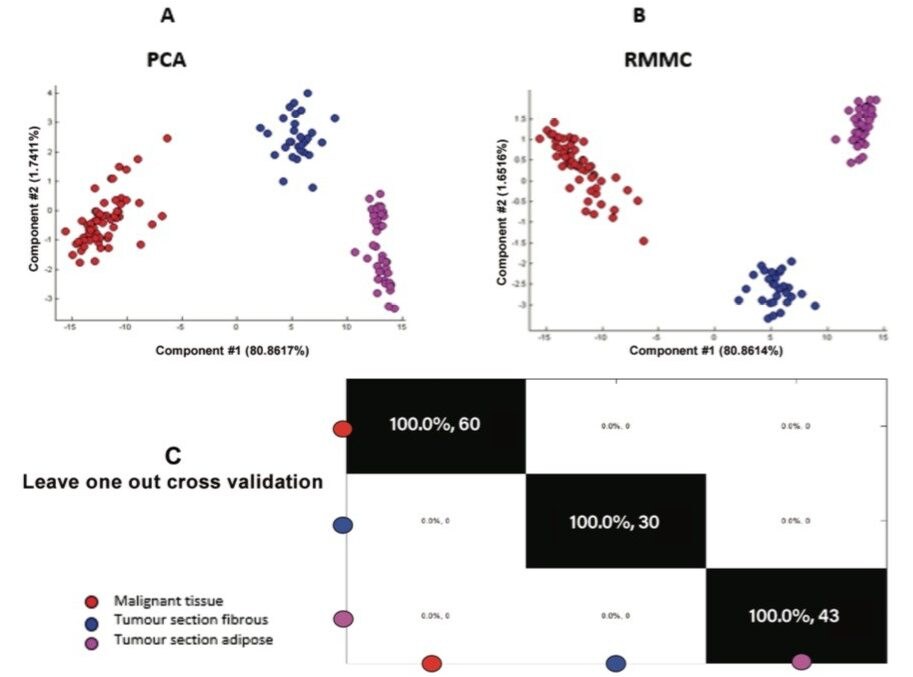
A range of other in-house or commercial statistical software packages are available. Another alternative investigated was to import raw imaging data into a MATLAB environment for further analysis. The individual grade 2 invasive ductal carcinoma (IDC) sample was subjected to unsupervised PCA to visualize differences between different regions within the section. Recursive maximum margin criterion (RMMC) analysis was used for supervised classification. Tissue types in each sample and their spatial distribution were determined by an independent histopathologist based on the H&E stained optical image (Figure 4). Based on this information, a small number of representative mass spectra per tissue were selected from the MS ion image to build a sample-specific RMMC model which was used to classify all pixels in the different tissue types. This data was submitted to cross validation, which achieved 100% accuracy in negative ion mode (Figure 5).
Desorption electrospray ionization mass spectrometry imaging can be effectively employed for spatial localization and distribution of molecules within breast tissue samples under ambient conditions for clinical research.
The advantages of DESI-MSI for the analysis of breast tissue samples include:
720005805, February 2017