
This application note discusses the work carried out to investigate the use of Waters UltraPerformance Convergence Chromatography (UPC2) and MS detection using the ACQUITY QDa as an alternative technology for the analysis of MAC (Methyl-3-aminocrotonate) without the prior need of derivatization, for the detection and quantification in an active pharmaceutical ingredient (API).
Mutagenic impurities, formerly known as genotoxic (GI) or potential genotoxic impurities (PGI), are compounds that have the potential to modify DNA, and as a consequence can cause cancer. It is important that impurities potentially present in the marketed drug are evaluated early in the drug development process. To that end, analytical methods must be developed that are sensitive and specific enough to determine the levels in both drug substance and product.
The International Conference on Harmonization published ICH M7 guidelines, which highlight the requirements for assessment and control of DNA-reactive impurities to ensure the safety of pharmaceutical products.1 The European Medicine Evaluation Agency (EMEA), U.S. FDA, and the Asia regulatory agencies all follow these guidelines. They require that any mutagenic impurities in a drug substance or drug product must be below the Threshold of Toxicological Concern (TTC) of 1.5 µg per day based upon the maximum daily dosage of the pharmaceutical compound over a lifetime. For example, for a dosage of 1 g of Active Pharmaceutical Ingredient (API) per day, any impurity must be less than 1.5 ppm (1.5 µg). This is orders of magnitude lower than for general pharmaceutical impurities analysis, which is at the 500 ppm level and governed by Q3B(R).2
Pharmaceutical analysis is typically performed using LC with UV detection for non-volatile compounds, or GC with FID detection for volatile compounds. However, the low levels of detection required for mutagenic impurities present a significant challenge. In these situations, MS detection is required in order to achieve the desired sensitivity. Some of these methods are required to provide support during the whole life cycle of a drug from early development through to manufacturing quality control. Typical reverse-phase (RP) (where the majority of separations are done on C18 stationary phases) and normal-phase (NP) chemistries can be used; this opens up a wide range of selectivity choices to help develop successful separations.
Convergence chromatography (CC) is a chromatographic technique similar to HPLC, but instead of the weak mobile phase being aqueous it is replaced with supercritical carbon dioxide (CO2). Supercritical CO2 can be paired with a large number of different co-solvents to increase the solvating power. CO2 is miscible with the whole range of the eluotropic series opening up a large choice of solvent selectivity – with methanol, IPA, ethanol, and acetonitrile being the most commonly used co-solvents.
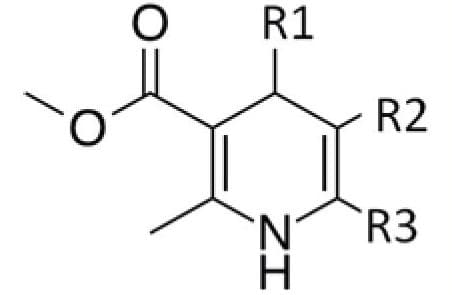
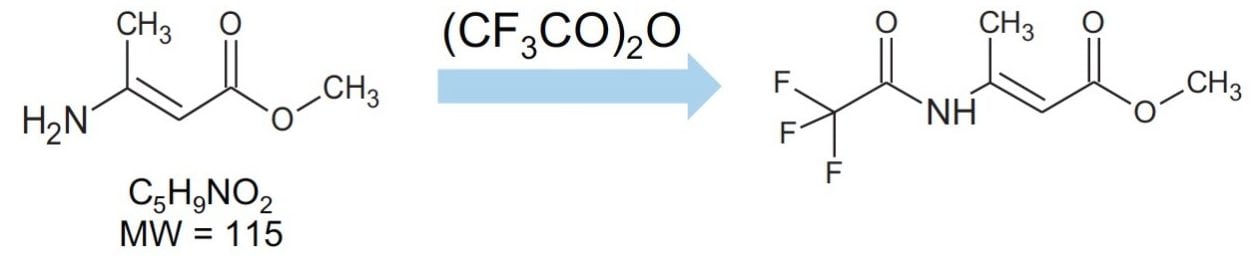
Methyl-3-aminocrotonate (MAC) is a Michael-reactive receptor and a starting material in a number of different cardiovascular drug products. The API used is an active substance from a proprietary drug product; therefore, only the partial structure is shown in Figure 1. MAC flags up a positive from the mutagenic structural alerts. This compound would typically be analyzed by static head space (SHS) GC-MS after derivatization with trifluoroacetic anhydride to increase the volatility (Figure 2).3 When MAC (underivatized) was analyzed by UPLC-MS nothing was seen; this was thought to be due to its poor stability in aqueous solvents.
In this type of trace analysis where there is a large amount of matrix it would be advantageous if chemical derivatization of the mutagenic impurity can be avoided for the following reasons:
To avoid the above issues, this application note discusses the work carried out to investigate the use of Waters UltraPerformance Convergence Chromatography (UPC2) and MS detection using the ACQUITY QDa as an alternative technology for the analysis of MAC without the prior need of derivatization, for the detection and quantification in an active pharmaceutical ingredient (API).
10, 5, 2.5, 1, and 0.5 ppm standards of MAC (with respect to 1 mg/mL API) were prepared in methanol.
|
System: |
ACQUITY UPC2 |
|
Column: |
Viridis BEH, 1.7 μm, 2.1 mm x 100 mm |
|
ABPR: |
1700 psi |
|
Column temp.: |
40 °C |
|
Sample temp.: |
15 °C |
|
Injection volume: |
1 μL |
|
Flow rate: |
1.5 mL/min |
|
Mobile phase A: |
CO2 |
|
Mobile phase B: |
MeOH |
|
Gradient: |
5% to 95% B at 1.5 mins, held until 2.1 mins then 5% B |
|
Run time: |
2.5 mins |
|
Make up solvent: |
MeOH, 2% H2O and 0.1% formic acid |
|
Make up flow: |
0.6 mL/min |
|
MS system: |
ACQUITY QDa Detector |
|
Ionization mode: |
ESI positive |
|
Single ion recording (SIR): |
m/z 116.1 Da [M+H]+ |
|
Capillary voltage: |
0.8 kV |
|
Sampling frequency: |
5 Hz |
|
Probe temp.: |
600 °C |
|
Cone voltage: |
8 V |
|
Data management: |
MassLynx Software v4.1 |
The structure of MAC is shown in Figure 2. It has a nominal molecular weight of 115 Da. Full scan analysis on the ACQUITY QDa Detector detected the expected [M+H]+ ion at m/z 116.1 Da.
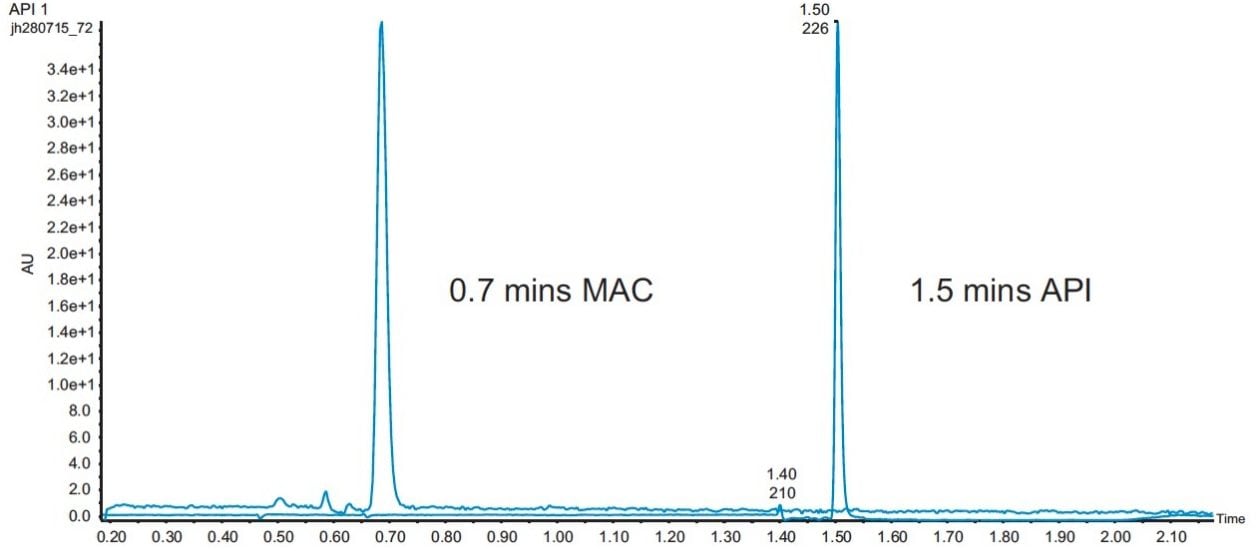
A number of different mobile B eluents were tried – including methanol, methanol with 0.1% formic acid, and methanol with 20 mM ammonium formate. The final method resulted in an elution time of 0.7 minutes for the MAC and 1.5 minutes for the API (Figure 3
ACQUITY QDa Detector probe temperature and cone voltage conditions were optimized for maximum sensitivity for the MAC analysis.
Selectivity issues can arise during trace analysis because the target analyte is at low levels in the presence of a large concentration of API, a counter ion, or – in the case of drug products – excipients. It is important when carrying out this type of analysis that a series of samples of API or drug product are spiked with the corresponding mutagenic impurity. This will indicate if there are any issues relating to stability, ion suppression, or enhancement effects. In this analysis, samples were prepared by spiking into the API a 1 ppm MAC standard, then analyzed. The result of this experiment showed that the areas for the unspiked and spiked standard were comparable. This implies the matrix does not have an effect on the MS response of this analysis. The areas of both spiked and unspiked samples are overlayed as shown in Figure 4.
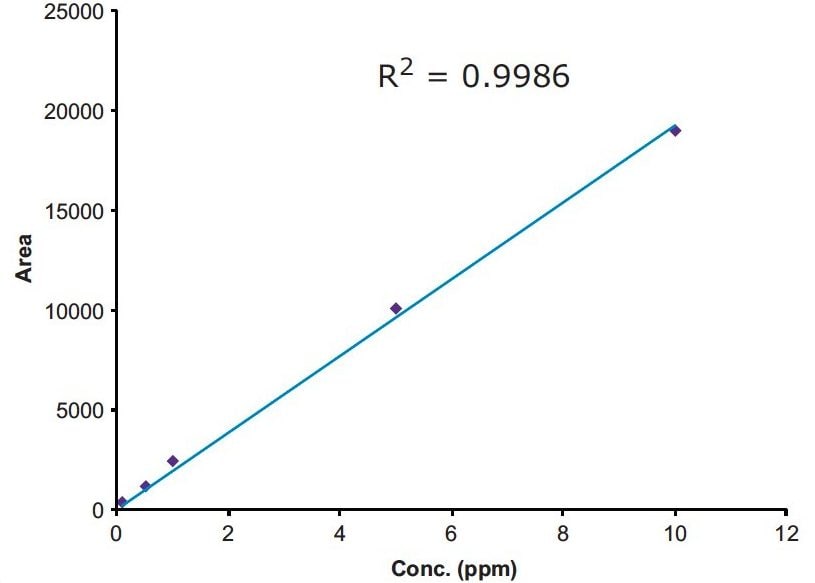
The linearity of the method was evaluated with five standards of 0.5, 1.0, 2.5, 5, and 10 ppm of MAC in methanol. The method showed good linear correlation between the peak areas and the ppm concentration with a correlation coefficient of R2 = 0.9985 (Figure 5). The signal-to-noise for the LOQ standard is more than 10 to 1 (Figure 6), and signal-to-noise ratio is 3 to 1 at the LOD standard (Figure 7). The percentage standard deviation of the six individual injections of all the five standards was less than 4%.

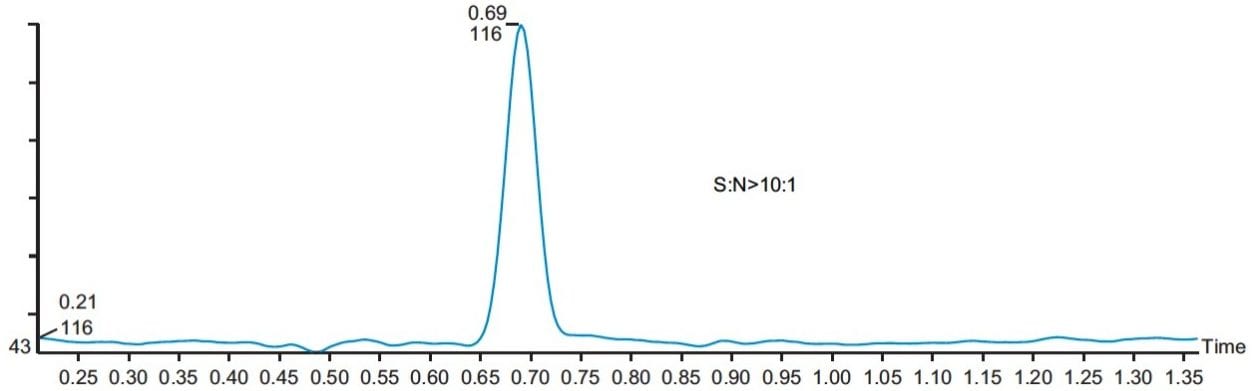
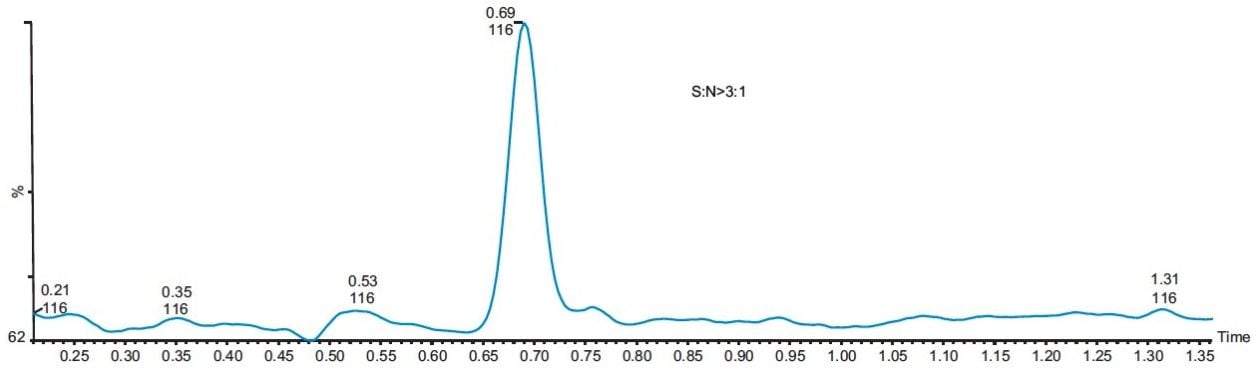
Three different batches of a 1 mg/mL solution of API in methanol were analyzed, and the results showed that they all contained less than 1.0 ppm of MAC. The overlay of a typical batch with a MAC 1 ppm standard is shown in Figure 8.
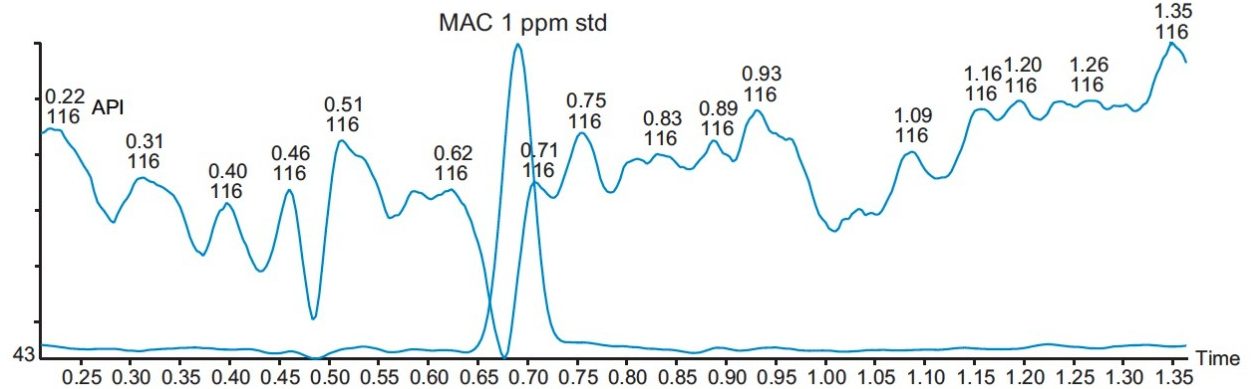
The ACQUITY UPC2 and ACQUITY QDa Detector with SIR provide a highly specific and sensitive method for the analysis of MAC down to a LOQ of 1.0 ppm related to 1 mg/mL API in solution
ACQUITY UPC2 and ACQUITY QDa Detector combination is an excellent opportunity for high-sensitivity trace analysis, and should be included as part of the toolkit for the analysis of mutagenic impurities
ACQUITY UPC2 and ACQUITY QDa Detector can be used through all stages of drug development and into a QC environment, if required
Fast analysis time because no derivatization and less validation was required
Alternative analysis for aqueous-sensitive components
720005846, November 2016