In this application note we propose an orthogonal method, UPLC HILIC/TUV-MS, in which the information about glycan heterogeneity and site occupancy is preserved. This method is complementary to UPLC HILIC/FLR analysis of the released glycans and the RP peptide map. The same tryptic digest used for the peptide map can be used in the method. The ACQUITY UPLC System with a UPLC BEH Glycan Column is used for UPLC/FLR analysis of released glycans and the proposed method for the separation of glycopeptides.
With this method, information about glycan heterogeneity and site occupancy is preserved and the same tryptic digest used for peptide mapping can be used. Since it does not require glycan release and labeling, complexity of sample preparation is reduced. This method is useful in the development and quality control of new protein-based therapies.
Glycosylation of proteins affects their tertiary structure and potentially therapeutic efficacy. Therefore, the glycosylation of therapeutic proteins such as monoclonal antibodies (mAb) needs to be closely monitored.
Reversed-phase liquid chromatography (RP-LC) is a primary method chosen for protein characterization via peptide mapping. Peptide mapping applications require efficient columns to resolve complex peptide mixtures into unique peptides. Modified peptides, such as oxidized or deamidated ones, can also be separated from the unmodified peptides.1 UltraPerformance Liquid Chromatography (UPLC) technology provides the resolving power needed for these challenging separations.2
It has been reported that RP-UPLC is able to resolve glycosylated peptides into their glycoforms.3 However, the complete resolution of glycopeptide micro-heterogeneity (same peptide sequence, various glycoforms) remains difficult. This is because retention in RP-LC is mainly due to peptide hydrophobicity, and is less affected by the presence of hydrophilic glycans. The separation is further complicated by the presence of non-glycosylated peptides in the sample that often elute in the vicinity of the glycopeptides of interest.
Several separation methods are available for glycan analysis, including capillary electrophoresis (CE), high pH anion exchange chromatography with pulsed amperometric detection (HPAEC-PAD), and hydrophilic interaction chromatography with fluorescent detection of labeled glycans (HILIC/FLR). While those methods are useful, the confirmation of glycan identity relies on their retention time, available standards, and use of specific exoglycosidase enzymes.4 Fraction collection of resolved glycans is often combined with a matrix assisted laser desorption/ionization (MALDI) MS method for confirmation of mass of glycans and their MS/MS structure identification. Because of the advantages of on-line MS, the off-line MALDI method is being recently replaced with LC-MS glycan analysis.
Two LC-MS methods currently under development are MS analysis of the intact proteins and LC/FLR-MS analysis of the glycan released from a glycoprotein. In the first case, the mass spectrum (after deconvolution) provides information about the protein molecular weight and its heterogeneity due to glycosylation.5 For mAbs, where the glycosylation nature is well understood, the intact mass information can be translated into the relative quantitation of glycoforms.6 Though useful as fast screening, the intact protein MS method may fail to detect minor glycoforms.
The second method for glycoprotein characterization utilizes specific enzymes (PNGase F) to release N-linked glycans from the protein. Glycans are typically enriched, labeled with fluorescent dye, and analyzed in HILIC mode. Highly efficient UPLC HILIC columns have been shown to facilitate an excellent glycan separation and relative quantification.7
HILIC separation of glycans is considered to be a reliable method. However, for proteins with multiple N-linked glycosylation sites, released glycans of the same type elute in chromatogram as cumulative peaks. Therefore, the information about the occupancy of different N-linked sites is lost. This is also the case for CE and HPAEC-PAD methods. While this does not present a problem for proteins with a single glycosylation site, such as monoclonal antibodies, it precludes full characterization of proteins with multiple glycosylation sites.
In this application note we propose an orthogonal method, UPLC HILIC/TUV-MS, in which the information about glycan heterogeneity and site occupancy is preserved. This method is complementary to UPLC HILIC/FLR analysis of the released glycans and the RP peptide map. The same tryptic digest used for the peptide map can be used in the method. The ACQUITY UPLC System with a UPLC BEH Glycan Column is used for UPLC/FLR analysis of released glycans and the proposed method7 for the separation of glycopeptides.3
|
LC system: |
Waters ACQUITY UPLC System |
|
Column: |
ACQUITY UPLC BEH Glycan Column, 2.1 x 150 mm, 1.7 μm amide sorbent |
|
Column temp.: |
40 °C |
|
Flow rate: |
0.2 mL/min |
|
Mobile phase A: |
10 mM ammonium formate, pH 4.5 |
|
Mobile phase B: |
10 mM ammonium formate, in 90:10 acetonitrile/water |
|
Gradient: |
90 to 55 % B in 45 min (81 to 49.5% acetonitrile in 45 min) |
|
Detection: |
ACQUITY UPLC TUV, 280 nm |
|
MS system: |
SYNAPT MS system |
|
MS acquisition: |
ESI positive ion V-mode, collision cell energy 5 V, 0.3 sec acquisition cycle, capillary voltage 3.0 kV, cone voltage 37 V, source temp. 100 °C, desolvation temp. 250 °C, cone gas 10 L/h, desolvation gas 550 L/h |
|
Sample: |
Humanized mAb tryptic digest |
Figure 1 illustrates the retention differences between peptides and glycopeptides in HILIC chromatography mode. The glycopeptides are highlighted with a blue box. The presence of highly polar glycan moiety greatly improves the retention of glycopeptide(s) of interest, and therefore they are well resolved from the remaining non-glycosylated tryptic peptides generated by the tryptic digest of the mAb. This specific retention behavior has been confirmed with other glycoprotein digests and appears to be a generic behavior of all glycopeptides, including the O-linked ones (data not shown).

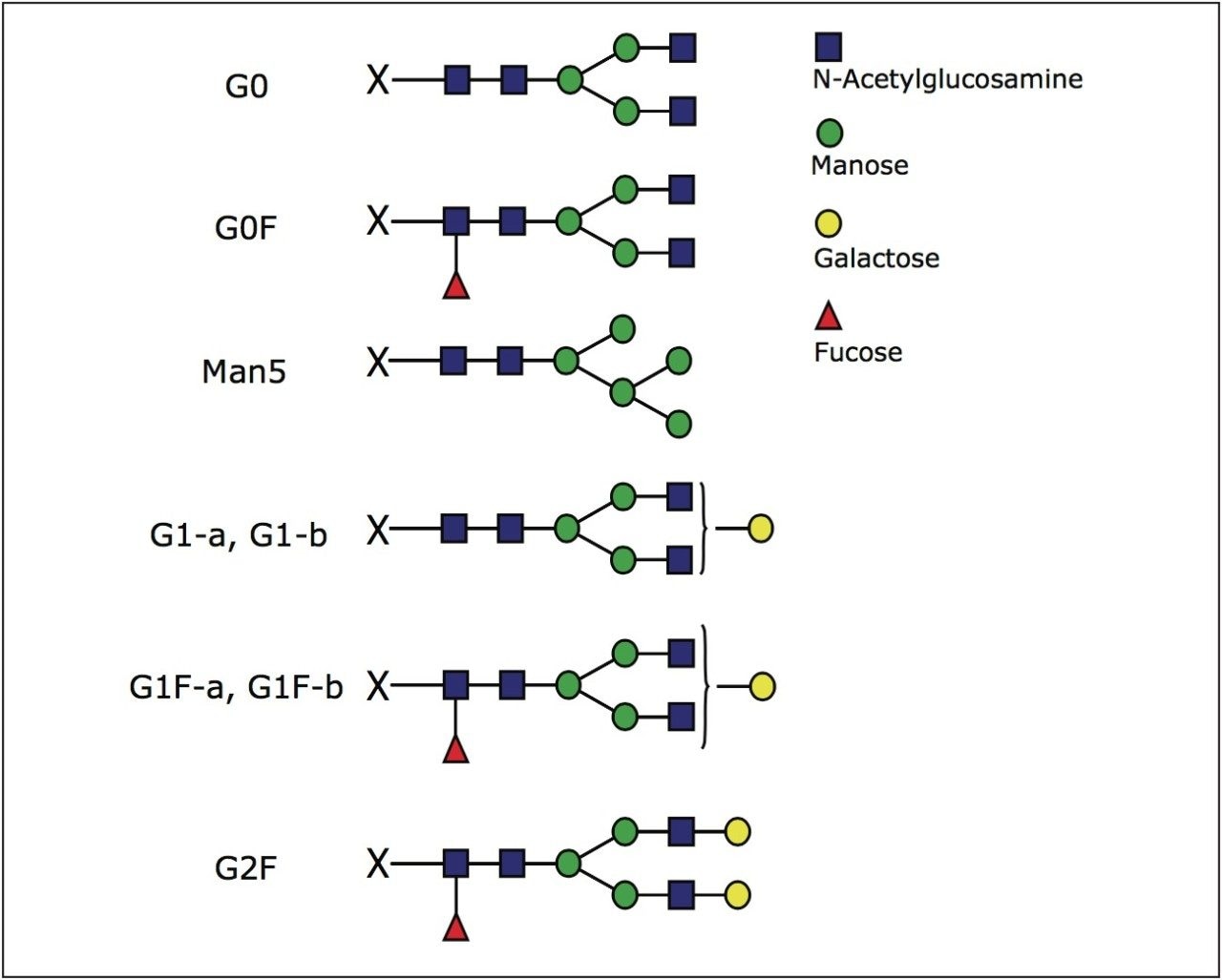
Further inspection of Figure 1 reveals that the glycopeptide EEQYNSTYR with glycans attached at asparagine position is resolved into seven distinct peaks. The pattern and intensity resembles the separation obtained for glycans released with PNGase F, labeled with 2-AB dye, and analyzed in HILIC mode using the same column (Figure 2). Glycan structures identified in this study are shown in Figure 3.

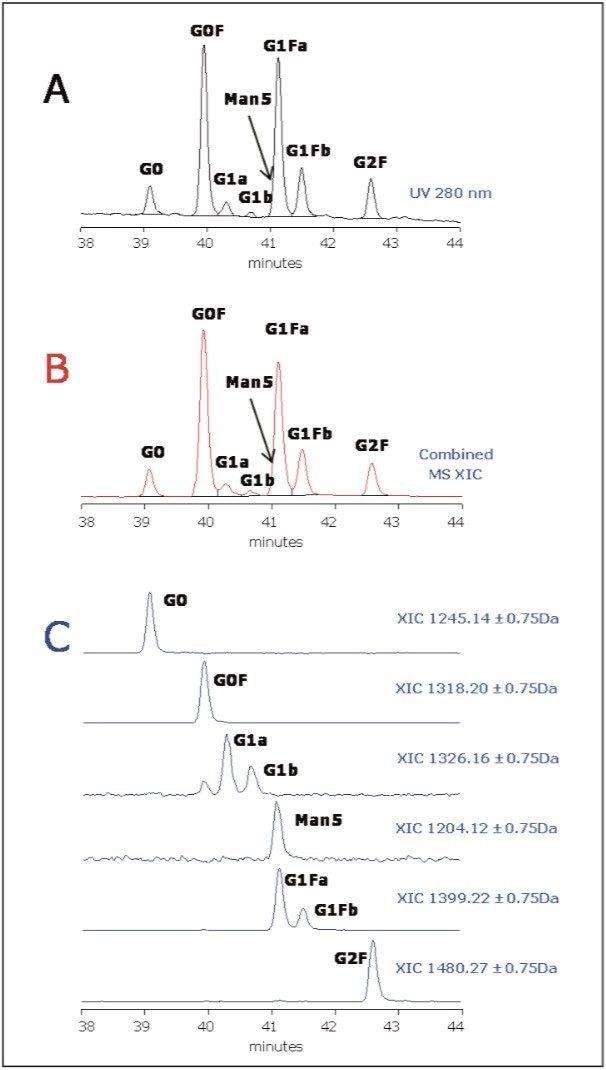
The expanded view of the glycoprotein separation highlighted in Figure 1 is presented in Figure 4. Seven main peaks are clearly seen in chromatogram.
Since the EEQYNSTYR peptide contains two aromatic amino acids, it absorbs UV light at 280 nm and serves as “tag,” providing equimolar response for all glycopeptides. Therefore, the relative quantitation of glycan micro-heterogeneity could be measured at 280 nm with little UV background interferences from the mobile phase.
Peak identity was confirmed by accurate MS data as shown in Figure 4C. When extracting XIC (extracted ion chromatograms) for expected glycan species in mAb, we detected the presence of G0, G0F, G1 G1F, and G2F glycans, and also Man5 variant that is coeluting with the dominant G1Fa peak (Figure 4C).
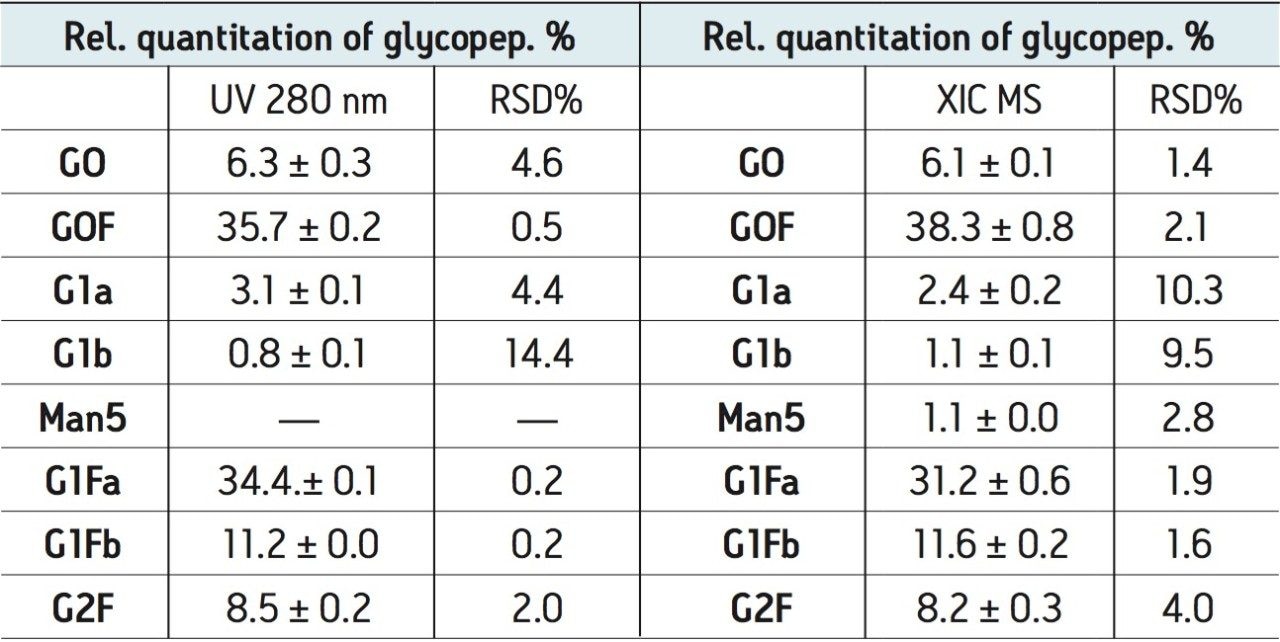
Relative quantitation based on UV 280 nm and XIC data was performed for three repetitive analyses. The results are listed in Table 1. Interestingly, the similar pattern in Figures 4B and 4C and similar quantitation results in Table 1 suggest that the glycopeptide variants have rather uniform MS responses (at least for neutral glycans observed in this study, see Table 1). MS also enables quantitation of glycopeptides that are not resolved and cannot be quantified by UV.
There are several benefits of this proposed orthogonal method compared to HILIC/FL analysis of released glycans in terms of easier sample preparation, use of the same sample as for peptide maps in RP, and information regarding the site occupancy of glycosylation site(s). In comparison with released glycan using HILIC with fluorescence detection is more sensitive than UV. Tens of pmole of protein digest or more needs to be injected on column for the proposed HILIC/UV glycopetide method, while HILIC/FLR requires only injection of sub pmol amounts of sample. Also, the resolution of released glycans in HILIC mode appears to be better than for glycopetides (compare Figures 2 and 4).
This application note describes a novel UPLC HILIC/TUV-MS method for characterization of protein glycosylation. Benefits of the new method include:
We believe that the proposed method is suitable for characterization of glycoproteins and in particular for monoclonal antibodies, which represent the main class of biotherapeutics. This method can speed up the development and quality control of new therapies, as well as improve safety and efficacy of protein drugs.
Preliminary results (not shown) suggest that the proposed method is useful also for characterization of O-linked glycans. Because of the lack of specific and efficient enzymes for their release, characterization of O-linked glycans in form of glycopeptides is a promising alternative that will be further investigated.
720003363, March 2010