For research use only. Not for use in diagnostic procedures.
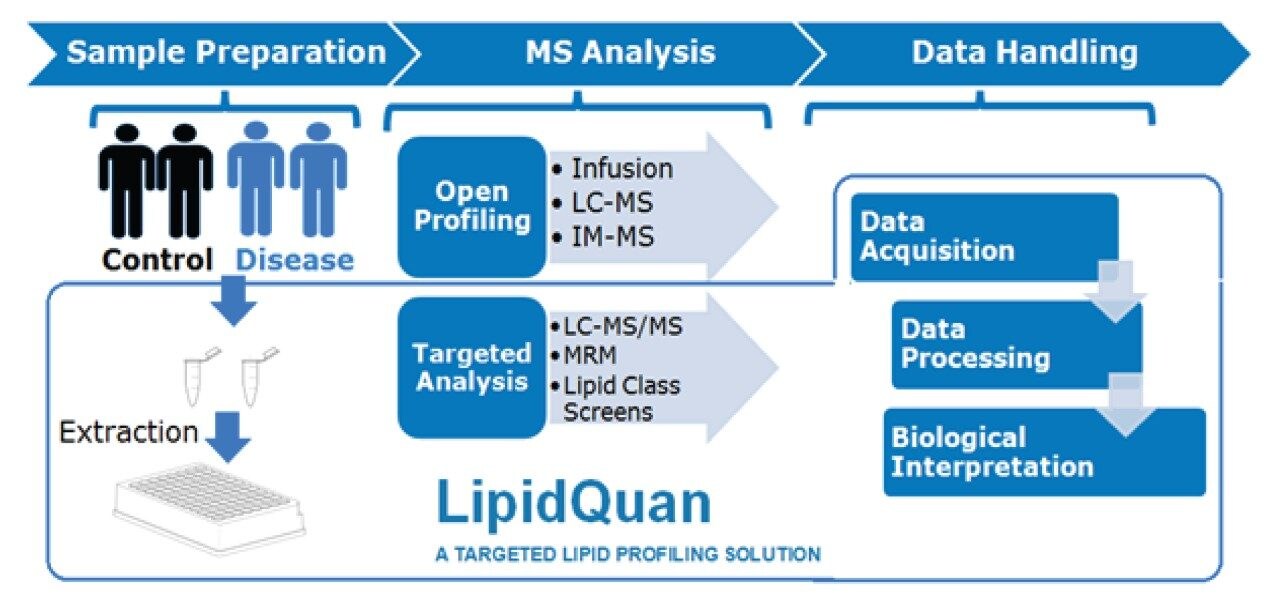
Although advances in mass spectrometry (MS) have allowed for more in-depth lipidomic analysis, unambiguous identification and quantification has proven difficult as lipids exhibit a high number of isomeric and isobaric species. In this application note, we adapt the LipidQuan workflow to provide a comprehensive and quantitative overview of the lipid species from murine adipose tissue extracts.
Leucine-rich-α2-glycoprotein 1 (LRG1) is a highly conserved member of a leucine-rich repeat grouping of proteins involved in protein-protein interaction, signalling, and cell adhesion.1 LRG1 is a secreted glycoprotein and was recently described as a novel angiogenic factor with a role in proliferative diabetic retinopathy.1 However, LRG1 is also expressed by adipose tissue, and has been suggested to be a liver and adipose specific secretory protein.2
Although advances in mass spectrometry (MS) have allowed for more in-depth lipidomic analysis, unambiguous identification and quantification has proven difficult as lipids exhibit a high number of isomeric and isobaric species. Here, we adapt the LipidQuan workflow3 (Figure 1) to provide a comprehensive and quantitative overview of the lipid species from murine adipose tissue extracts.
Adipose tissue (~30 mg) from wild type and transgenic LRG1 knockout mice (LRG1tm1(KOMP)Vlcg) were extracted using a chloroform:methanol (2:1) Bligh and Dyer method4 and subsequently dried down for transportation and analysis.
Samples were reconstituted in 500 μL isopropanol (IPA). Fifty microliters (50 μL) of each sample were pooled in a glass vial (600 μL total volume) and were spiked with stable isotope labelled (SIL) standards (SPLASH LIPIDOMIX and deuterated Ceramide LIPIDOMIX, Avanti Lipids, Alabaster, AL) at seven concentration levels to generate calibration curves for quantification. High, middle, and low QC samples at 80%, 8%, and 1.275% of the highest concentration of the calibration curve were used.
Six biological replicates representing wild type as controls and six mutant mice were analyzed. Samples were injected twice for both positive and negative mode analyses.
A simple sample preparation procedure was adopted using 40 μL of extract reconstituted in IPA spiked with 5 μL of Odd Chain LIPIDOMIX (1:100, Std. Mix:IPA) and 5 μL of neat IPA in total recovery Waters UPLC vials (p/n: 186005669CV). Samples were vortex mixed prior to LC-MS/MS analysis.
|
LC system: |
ACQUITY UPLC I-Class PLUS or H-Class PLUS System (Fixed Loop (FL) or Flow Through Needle (FTN)) |
|
Column(s): |
ACQUITY UPLC BEH Amide 2.1 × 100 mm, 1.7 μm |
|
Column temp.: |
45 °C |
|
Flow rate: |
0.6 mL/min |
|
Mobile phase: |
95:5 Acetonitrile/water + 10 mM Ammonium acetate (A) and 50:50 Acetonitrile/water + 10 mM Ammonium acetate (B) |
|
Gradient: |
0.1% to 20.0% B for two minutes, then 20% to 80% B for three minutes, followed by three minutes re-equilibration |
|
Run time: |
Eight minutes |
|
Injection volume: |
2 μL |
|
MS systems: |
Xevo TQ-XS, TQ-S, or TQ-S micro |
|
Ionization mode: |
ESI (+/-) |
|
Capillary voltage: |
2.8 kV (+) 1.9 kV (-) |
|
Acquisition mode: |
MRM |
|
Source temp.: |
120 °C |
|
Desolvation temp.: |
500 °C |
|
Cone gas flow: |
150 L/hr |
|
Desolvation flow: |
1000 L/hr |
|
Nebulizer gas: |
7 bar |
|
Ion guide offset 1: |
3 V |
|
Ion guide offset 2: |
0.3 V |
The LipidQuan Comprehensive Method Package containing the LC conditions, MS method, and associated TargetLynx processing method (including retention times and MRM transitions) was downloaded from the Waters Targeted Omics Method Library (TOML) and used to generate these methods. The resulting data were processed using either TargetLynx or Skyline (MacCoss Lab Software, University of Washington). Statistical analysis was performed using SIMCA P+ (Umetrics, Umeå, Sweden) and further data interrogation conducted using Metaboanalyst.5
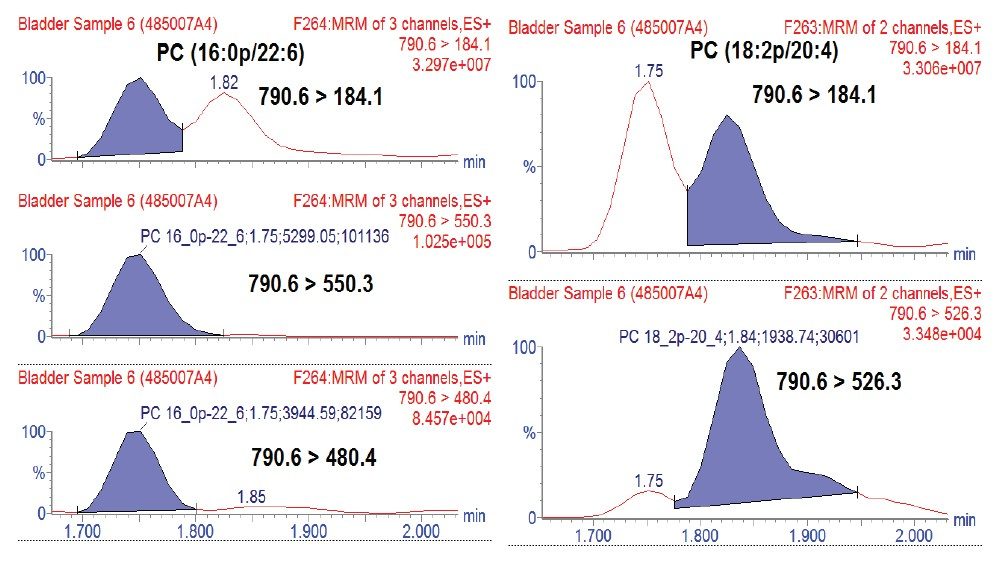
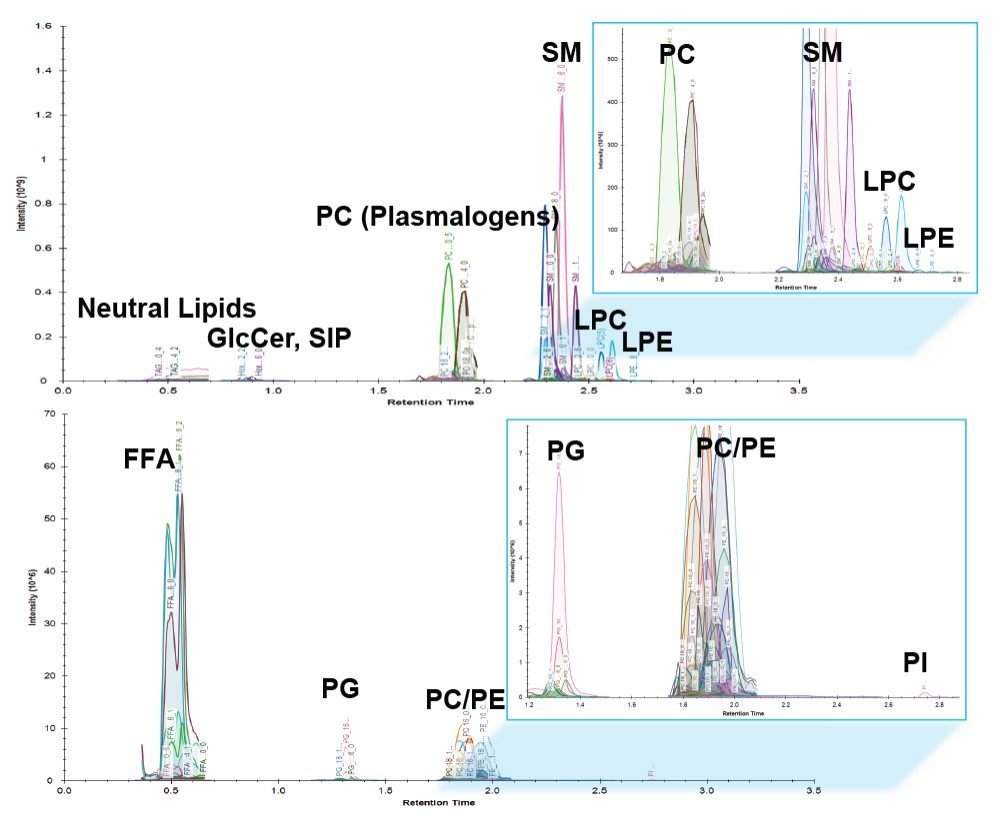
Lipid analysis of murine adipose tissue from wild type and transgenic LRG1 knock-out subjects were conducted using the LipidQuan platform. The samples were prepared for LC-MS/MS by re-constituting total extracts in isopropanol. Samples were randomized and two technical replicates per sample acquired. Targeted LC-MS/MS data were acquired in positive and negative ion electrospray modes using the LipidQuan method package that was downloaded from the TOML website. Deployment of LipidQuan method files using Quanpedia allowed for the fast and easy importing of all MRM transitions and chromatographic conditions, representing a positive and negative mode screen for lipid species. This simple workflow eliminated the manual input of LC-MS methods and eliminated possible transcription errors. The MRMs contain highly specific fatty acyl transitions, in addition to head group fragments for increased identification confidence (Figure 2). Example chromatograms of samples in both positive and negative mode are shown in Figure 3. Using this methodology, more than 400 lipids were identified in the samples.
Statistical analysis of the data revealed clear separation between the two sample cohorts. A PLSDA model created using SIMCA P+ and MetaboAnalyst resulted in clustering of wild types and mutant mice samples (Figure 4). The Q2 values were much greater than 40% (~90%), showing the model created is predictive. Permutation tests were also used to validate these models.
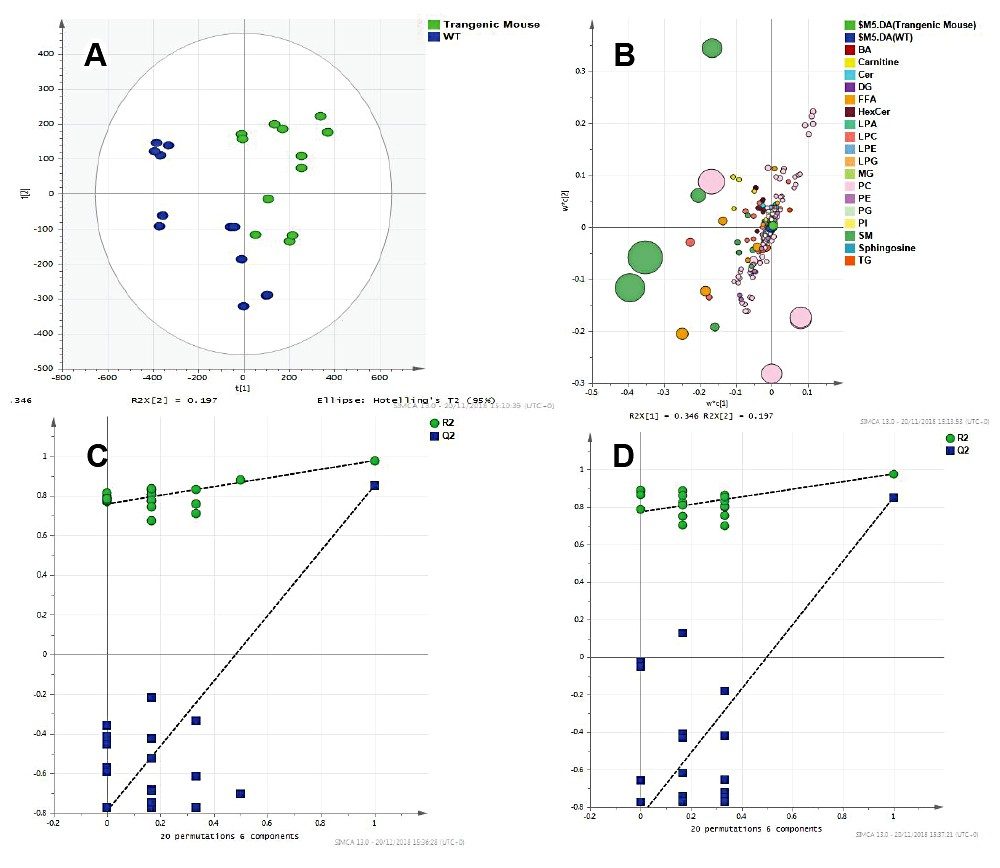
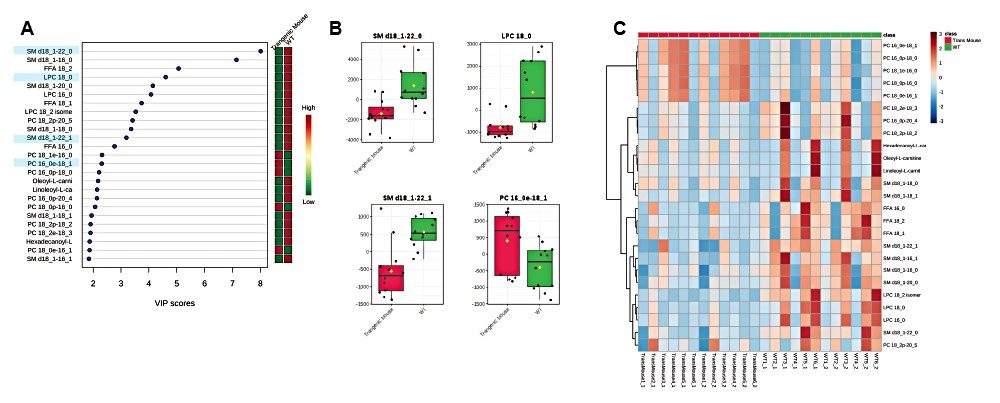
A VIP scores plot and heatmap based on VIP scores from the PLS-DA models (Figure 5) was generated using MetaboAnalyst. The top 25 VIP scores revealed LPC, PC, and SM classes to be the main contributors to sample type clustering. The majority of lipids have elevated levels in wild type mouse samples. However, some plasmalogens levels are elevated in transgenic mouse samples, for example PC (18:1e/16:0), PC (16e/18/1), and PC (18:0e16:1). The ANOVA/t-test, with %1 FDR, highlights lipid species with p-values greater than 0.05 and example chromatograms of these statistically significant peaks are overlaid to show potential for quantification (Figure 6).
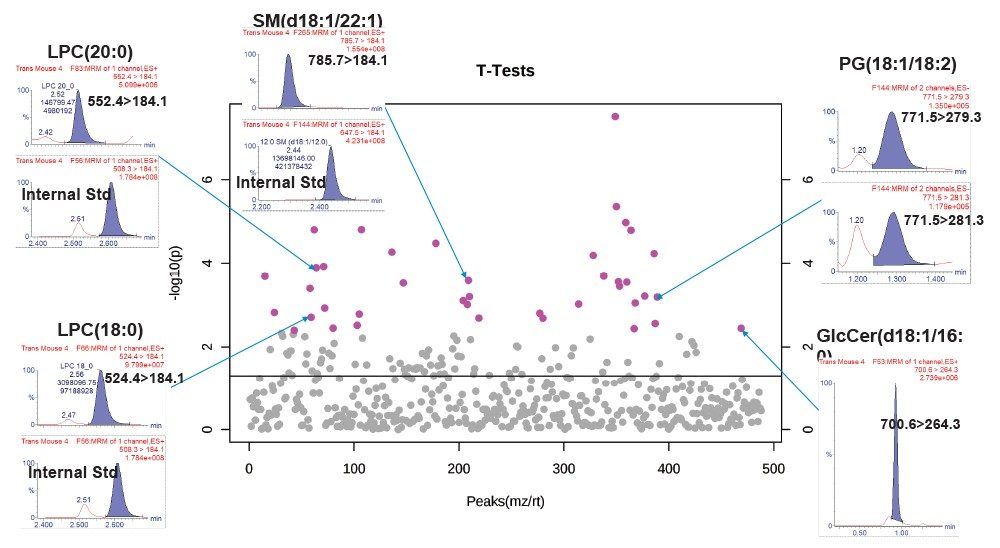
Quantification was achieved using calibration curves of spiked extracts with known concentrations of SIL standards prior to extraction. By using surrogate standards prepared and analyzed under identical conditions to those of endogenous lipids, the quantification of endogenous lipids within the same class was achieved. One caveat with regards to this method of quantitation is that the MS detector responses for lipid species from the same class are reported to vary in intensity depending on the FA chain length.6 The use of a commercially available, premixed SIL solution, rather than an SIL for each measured lipid, significantly reduces the overall cost of large studies. Deuterated standards were used to assess linear response, with typical R2 values ranging from 0.97–0.99 for the various lipid classes in both modes of ionization. Figure 7 shows example calibration curves and calculated concentrations for various lipids found to be statistically significant.
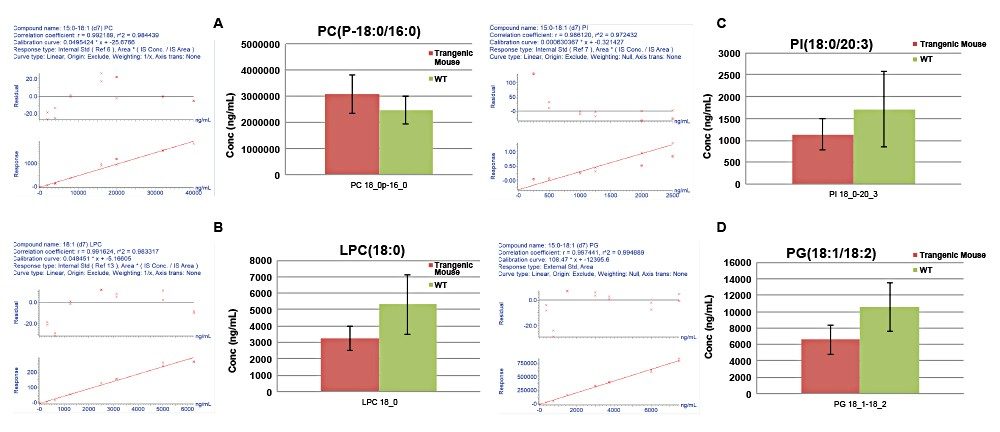
The genetic deletion of the putative adipokine, LRG1, appears to remodel membrane associated phospholipids in the brown adipose tissue. LRG1 -/- mice have enriched phosphatidylcholines containing abundant and monounsaturated fatty acyl chains (16:0/18:1) and depleted sphingomyelin species. A decrease in free fatty acid and L-carnitine species reflected alterations to energy metabolism.
720006758, February 2020