For research use only. Not for use in diagnostic procedures.
This is an Application Brief and does not contain a detailed Experimental section.
Thia Application brief demonstrates the utility of a larger scale chromatography system for plasma proteomics over traditionally employed systems. In addition to increased throughput, this methodology would be aligned with other omics analytical scale methods, such as metabolomics and lipidomics.
Identification and quantification of plasma based proteins utilizing the Biognosys PQ500 reference peptide kit on the ACQUITY UPLC I-Class System and 1 mm scale chromatography column.
Plasma proteomics, in a clinical research environment, requires samples to be analyzed with high throughput, utilizing an extremely reproducible chromatography system coupled to a mass spectrometer with the ability to produce data of high specificity. Quantitative proteomics often incorporates the use of stable isotope labelled (SILs) peptides spiked into the analyte in order to provide absolute quantitation. For use in plasma proteomics, there has been the recent introduction of Biognosys PQ500, a panel set of more than 500 SIL peptides. Normally, proteomic experiments involve very low sample amounts and require nanoscale chromatography to provide ultimate sensitivity. However, since the type of samples analyzed in plasma proteomics tend to have tryptic peptides present in relatively large amounts, previous experiments have involved the use of 300 micron scale chromatography.¹ In the experiments described here, we have scaled the chromatography further and investigated the use of the ACQUITY UPLC I-Class System equipped with a 1 mm I.D. column for sample introduction. In addition to increased throughput, this methodology provides increased flexibility and therefore provides the prospect of using a single platform for plasma proteomics, metabolomics, and lipidomics.
The samples analyzed consisted of tryptically digested, undepleted human plasma from three different conditions spiked with Biognosys PQ500 SIL peptides. Sample groups consisted of controls (n=6), chronic obstructive pulmonary disease (n=6), and asthma (n=6). Individual samples per group were pooled to provide three working samples. In addition, a pool of all conditions/samples acted as a QC. An ACQUITY UPLC I-Class System was equipped with an ACQUITY UPLC CSH130 C18, 1 mm x 100 mm long analytical column (p/n 186006934). Samples were analyzed in triplicate (QC = 5 injections) at two different loadings of 5 ug and 10 ug, which incorporated the PQ500 at ‘Injection Equivalents’ of one and two respectively. The samples were separated using a reversed phase gradient from 1% to 40% acetonitrile (+0.1% formic acid) over 15, 30, or 45 minutes at a flow rate of 50 µL/min. In order to minimize any post column peak broadening, a short piece of 60 micron I.D. PEEK tubing connected the output of the column to a low flow ESI probe. The mass spectrometer acquisition mode employed was SONAR DIA, which has been described elsewhere.² The same conditions were applied here. Data were processed using Progenesis QI for Proteomics and Spectronaut Pulsar X (Biognosys, Schlieren, Switzerland). Searches were performed using carbamidomethyl C (fixed) and oxidation of methionine (variable) modifications in addition to a 1% FDR.
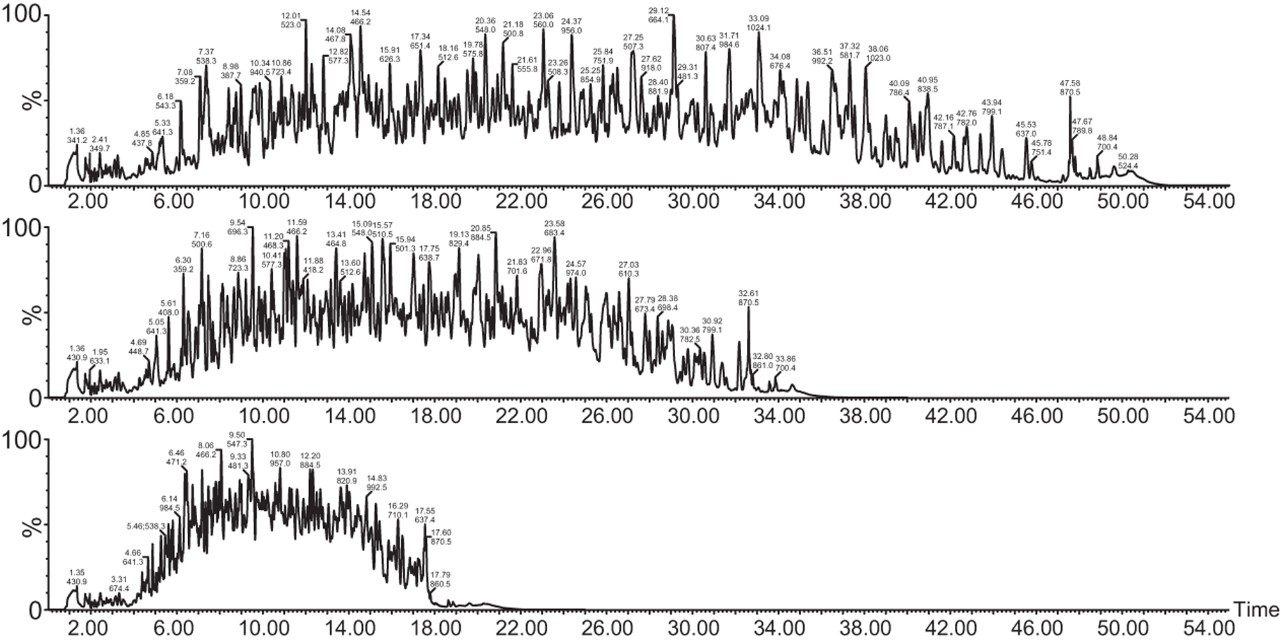
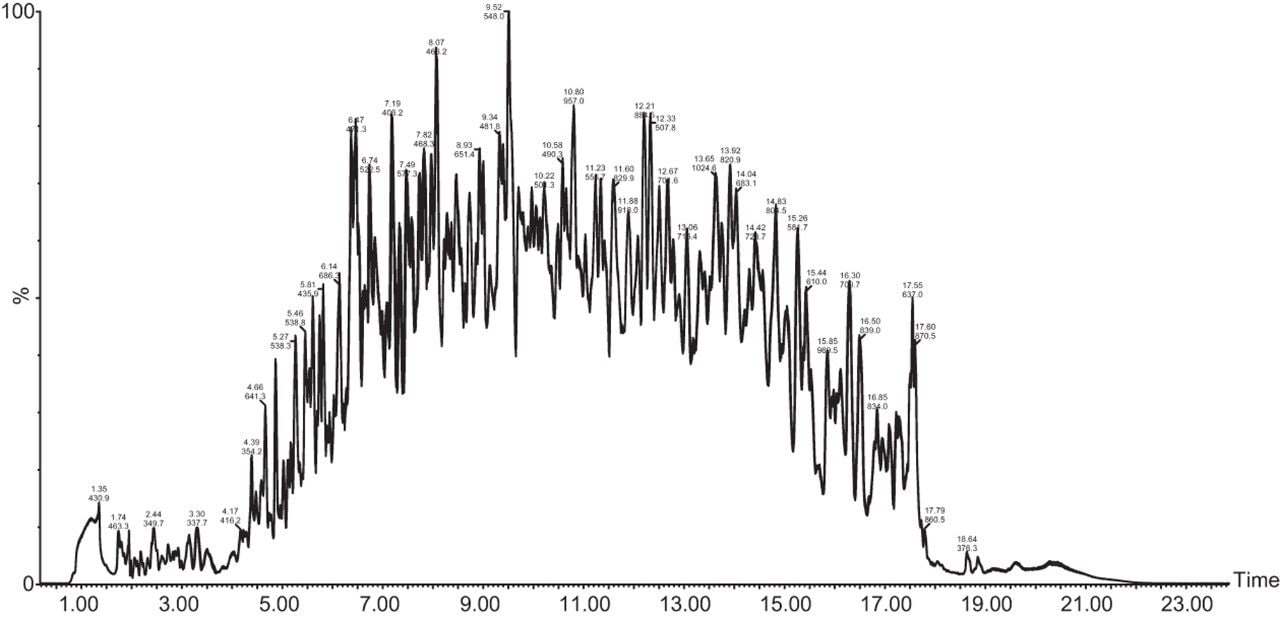
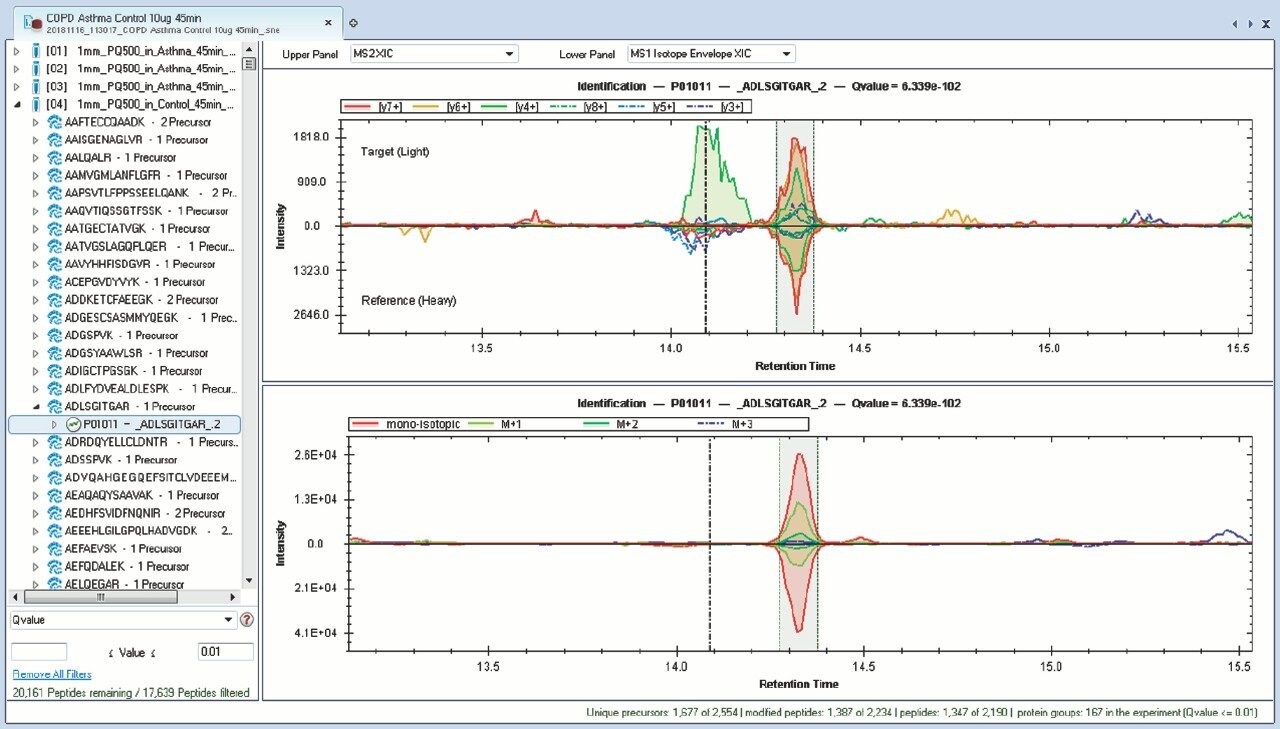
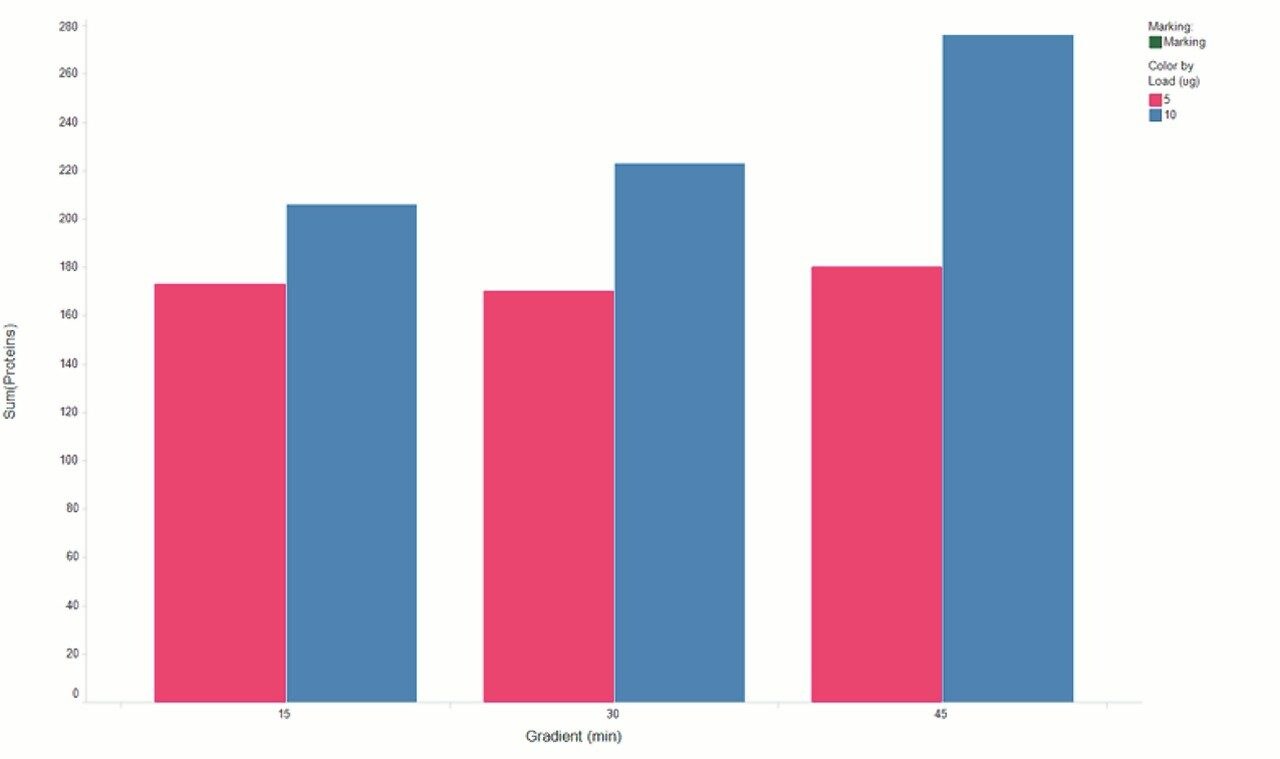
Chromatograms in Figure 1(a) represent the effect of gradient length on peptide separation, where shown are data from the three different gradient lengths and a load of 10 ug. The excellent chromatographic reproducibility obtained for the shortest gradient length is shown in Figure 1(b), which represents three overlaid chromatograms for 15 min gradient length and a 10 ug load. When data is processed using Spectronaut, heavy and light analogues are identified from the peptide panel. For a 10 ug load and 45 minute gradient, one such heavy and light identification is shown in Figure 2 for the peptide EDVYVVGTVLR from C4b-binding protein alpha chain (Uniprot accession: P04003). In total, 159 light/heavy pairs are identified in this experiment. The total number of quantified protein identifications from searches performed within Progenesis QI for Proteomics are shown in Figure 3. Gradient length had little effect on protein IDs for the 5 ug loadings but longer gradients were more beneficial for the 10 ug loading. The samples used for this study consisted of two different respiratory disease states, normal control, and a pooled QC. The PCA plot shown in Figure 4 indicates that separation of the different groups is readily achieved and corresponding CVs of <8% (Figure 5) further demonstrate the technical reproducibility of the data.
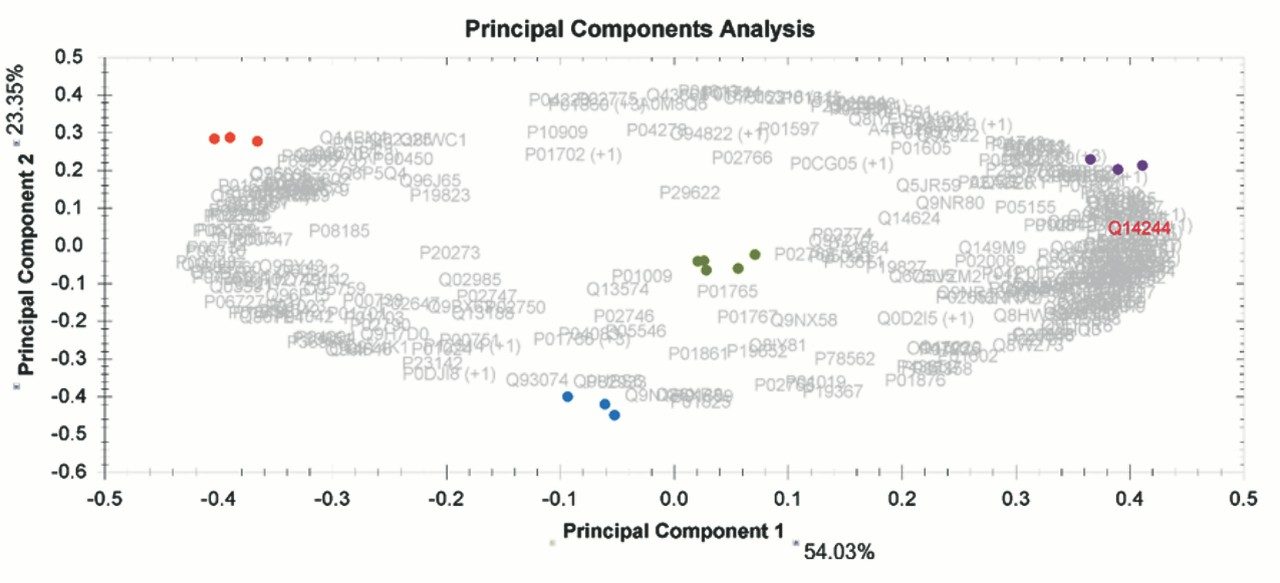
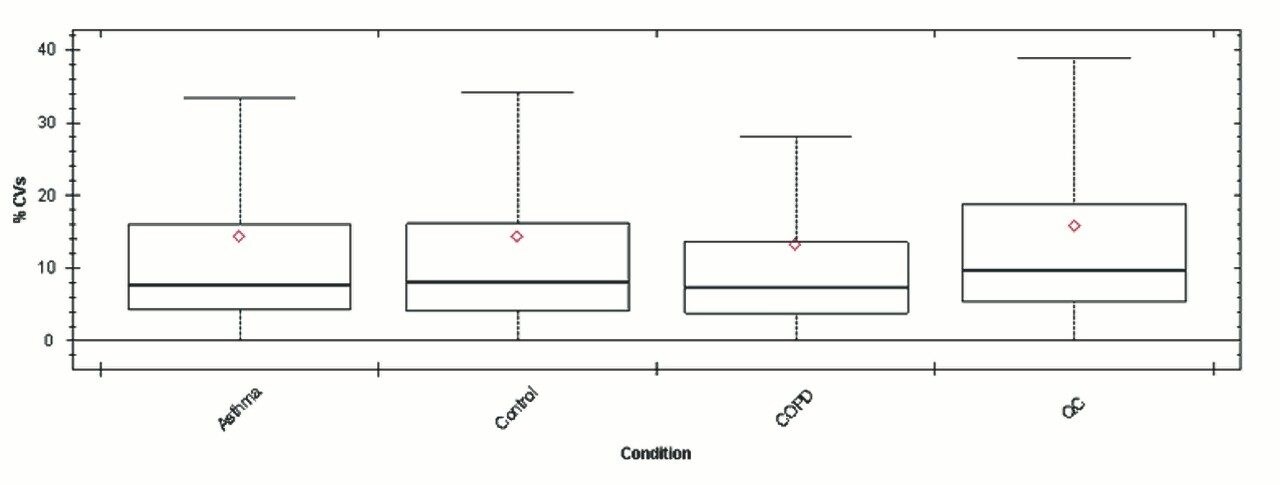
We have shown the utility of 1 mm scale chromatography for the analysis of typical samples from plasma proteomic experiments, where sample amounts tend to be present at elevated levels. The gradient has been delivered using an ACQUITY UPLC I-Class System and therefore raises the prospect of a single LC-MS platform for proteomics and other omic applications, such as metabolomics and lipidomics. The SONAR DIA acquisition method offers a compatible acquisition speed for rapid chromatographic methods of this type.
720006493, March 2019