This application note demonstrates the ability of a single method on the UPLC-MS system to perform routine analysis and quality control of small interfering RNA molecules. This method’s exceptional dynamic range will enable the low-level quantitation of modifications, process impurities, or contaminants in the presence of the main RNAi molecule facilitating regulatory requirements compliance.
The discovery of RNA interference mechanisms, now broadly used for silencing the expression of target genes, has created a new category of synthetic RNA oligonucleotides containing a matrix of modifications. The primary intent of these modifications to native RNA molecules is to increase binding constants, increase nuclease resistance, or help preserve unique secondary structure.
Because these molecules are synthetically produced step-wise in a solid phase process, the final product may contain multiple truncated oligonucleotides and a mixture of process-related impurities. It is critical to not only be able to detect modifications, process impurities, or contaminants, but to quantitate them as well since they may directly affect compound efficacy and safety.
Similar to any pharmaceutical product, these molecules must also be assayed for identity and purity.
In order to accomplish full characterization of these complex molecules, multiple analysis techniques are typically used, requiring a number of different types of instruments and highly-specialized laboratory technicians. To satisfy the need for a single, sensitive, quantitative, and high-throughput method for RNAi analysis, a method has been developed utilizing an ACQUITY UltraPerformance LC (UPLC) System, Oligonucleotide Separation Technology (OST) Columns and the Q-Tof Premier Mass Spectrometer for detection.
The Waters ACQUITY (UPLC) System, combined with ACQUITY UPLC OST Columns packed with 1.7 μm sorbent, offers superior analytical performance for oligonucleotide analysis compared to HPLC, fast LC separations, and other techniques.
|
LC system: |
Waters ACQUITY UPLC System |
|
Column: |
ACQUITY UPLC OST C18 2.1 x 50 mm, 1.7 μm |
|
Column temp.: |
60 °C |
|
Flow rate: |
0.2 mL/min |
|
Mobile phase A: |
15 mM TEA. 400 mM HFIP |
|
Mobile phase B: |
50% A, 50% methanol |
|
Gradient: |
20 to 40% B in 10 min |
|
Detection: |
ACQUITY UPLC PDA, 260nm |
|
MS System: |
Waters Q-Tof Premier Mass Spectrometer |
|
Capillary: |
2500 V |
|
Sample cone: |
35 V |
|
Extraction cone: |
3 V |
|
Ion guide: |
2.5 V |
|
Desolvation temp.: |
200 °C |
|
Source temp.: |
120 °C |
|
Cone gas flow: |
50 L/hr |
|
Desolvation gas flow: |
600 L/hr |
RNAi 21 nt (nucleotides), 5' -UUC UGU AAU CUC UUG UCU ATT -3', and 20 nt, 5' -UC UGU AAU CUC UUG UCU ATT -3', were purchased from Integrated DNA Technologies, Coralville, IA, U.S. The samples were reconstituted in 0.1 M triethylamine acetate (TEAA) to make 40 pmole/μL concentration for LC/MS analysis.
Exceptional resolution of the UPLC separation was achieved for the RNAi sample within 10 minutes, as shown in Figure 1. The full-length synthetic RNAi product was successfully resolved from its failure sequences.
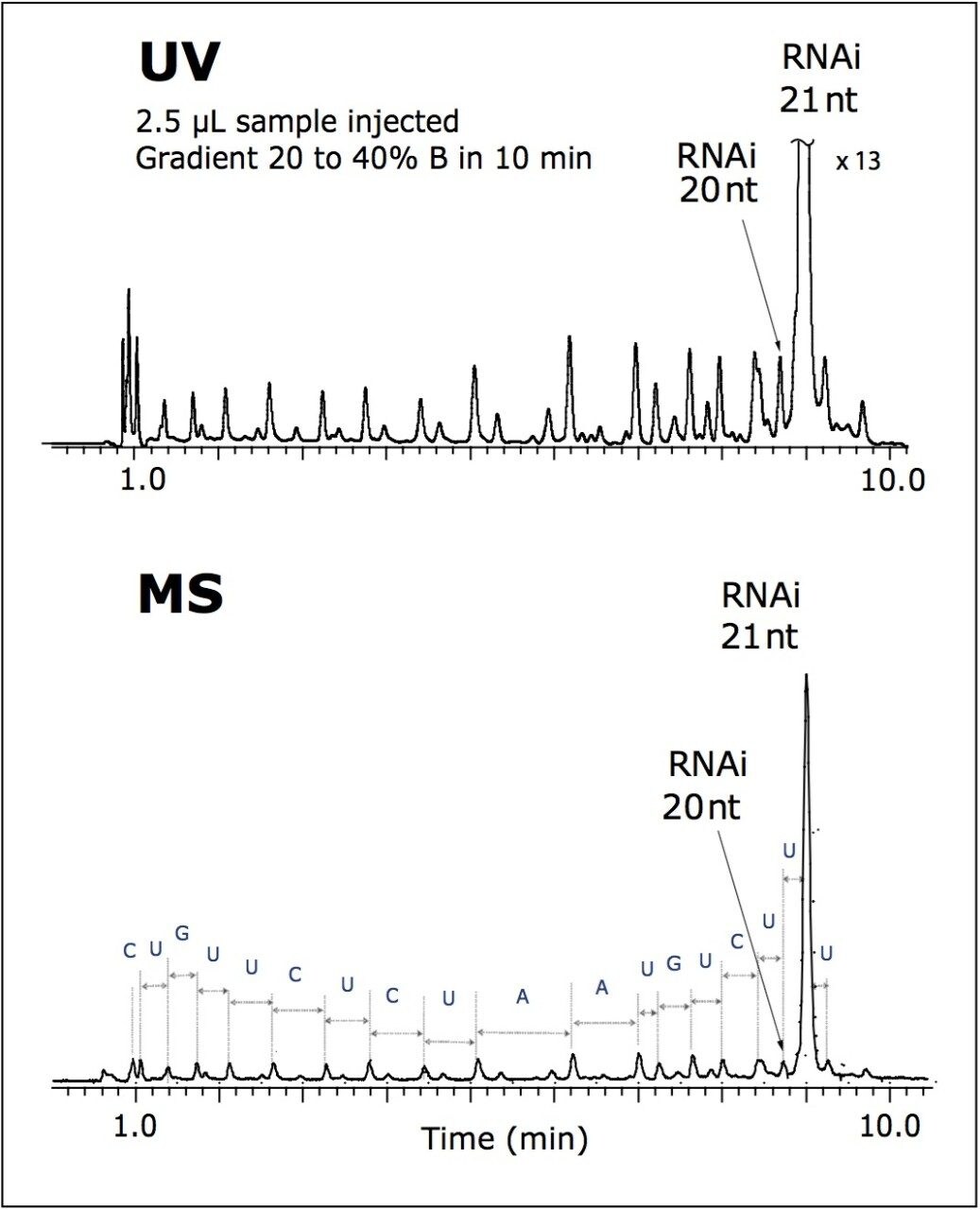
This method is suitable for the oligonucleotide purity determination and monitoring of the chemical synthesis efficiency.
Mobile phases containing 15 mM triethylamine (TEA), 400 mM hexafluoroisopropanol (HFIP), pH 7.9, and methanol are compatible with MS electrospray ionization. Choosing the narrow (75 μm I.D.) silica capillary tubing between the PDA detector and the ESI source decreased a post-UV void volume and reduced the broadening of the chromatographic peaks prior to MS detection.
The acquisition of the accurate masses allowed for an assignment of the peaks of 5' -truncated oligomers (failed sequences generated during oligo synthesis), as well as some other impurities. The mass of each peak in the MS chromatogram was deconvoluted using MaxEnt1 software.
The tentative 5' -end failure products are assigned in Figure 2. Nearly the entire sequence of the parent oligonucleotide was elucidated. MS analysis also revealed a presence of an extra uridine mononucleotide added to the target 21-mer RNAi sequence.
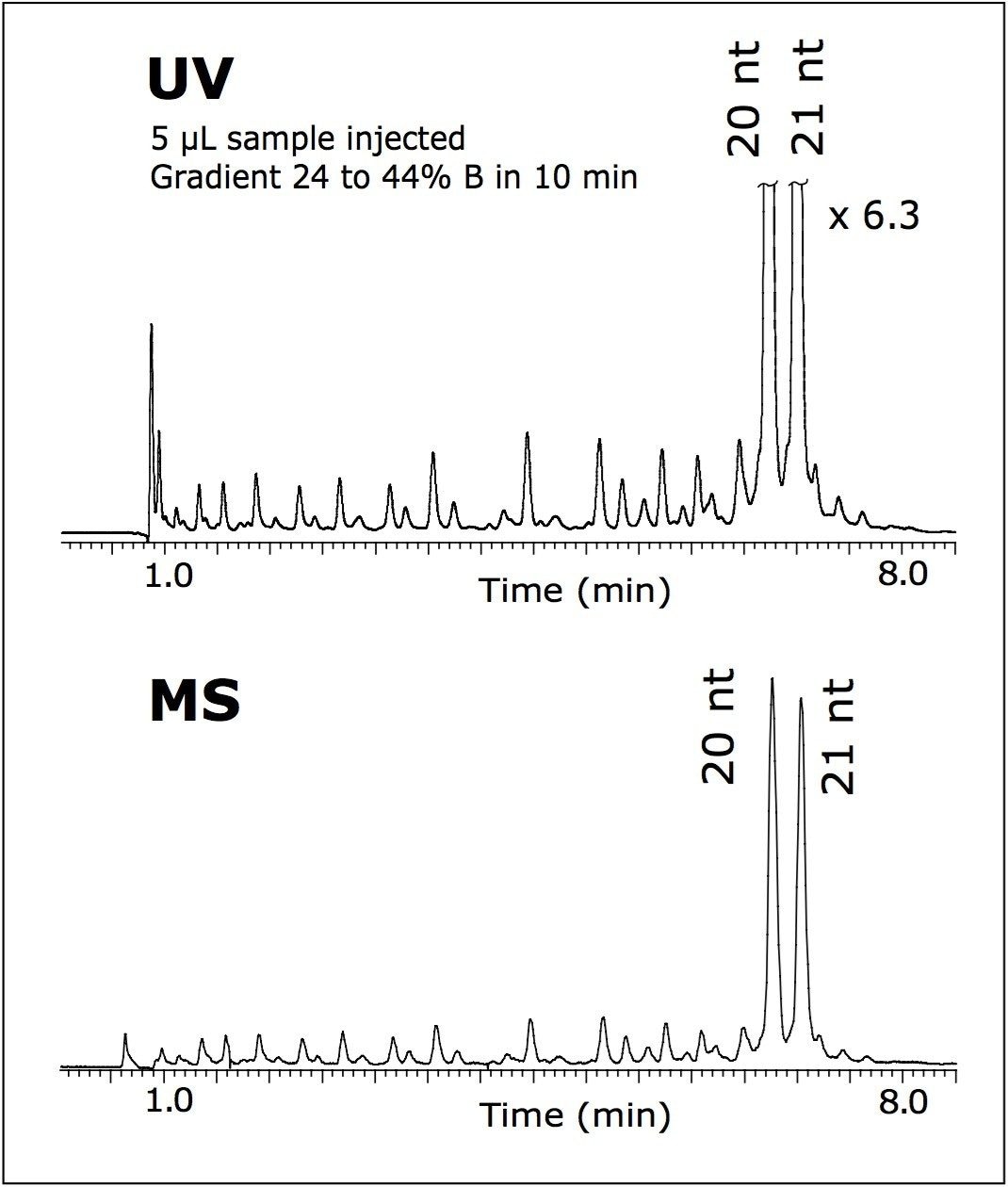
In order to verify the origin of the peak adjacent to the target 21 nt oligomer, an original sample was spiked with its 20 nt (N-1) homolog. Both oligonucleotides were mixed 1:1 to obtain 40 pmole/μL of each oligomer in a vial; 5 μL of the sample was injected on column.
The high efficiency of the developed method allowed the separation to resolve 21 nt from 20 nt N-1 RNAi within an 8-minute analysis (Figure 3). This experiment confirmed the correct assignment of the 20 nt peak as a failed sequence peak in Figure 1. Faster separation, shown in Figure 3, was achieved by adjusting the initial gradient strength as noted. When maintaining the same gradient slope, resolution is not negatively affected.
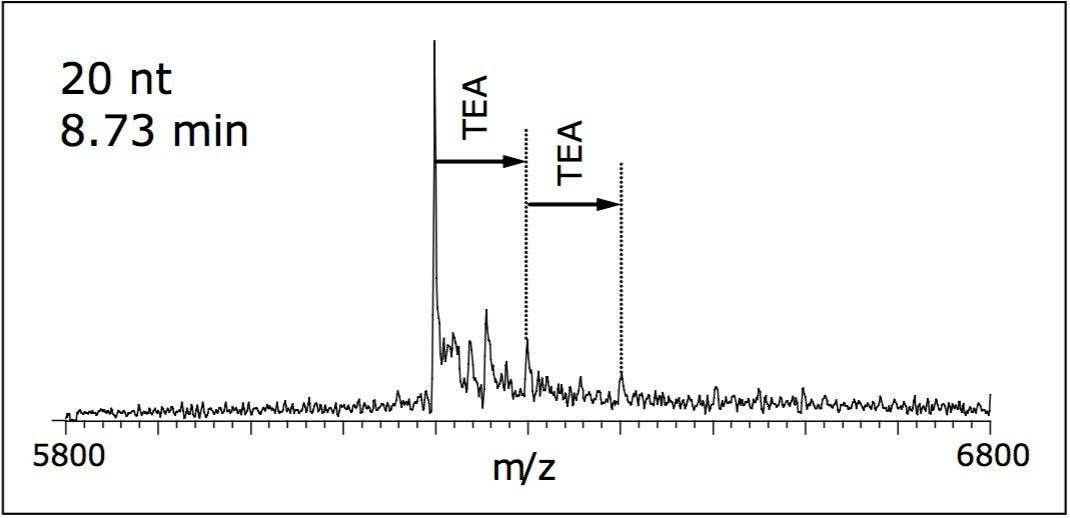
Formation of clusters and adducts from a buffer containing TEA was detected as 101 Da and 202 Da adducts in the deconvoluted mass spectra (Figure 2). Few sodium and potassium adducts were observed and did not obscure the mass spectral interpretation. MS parameters, including desolvation temperature, desolvation gas flow, cone gas flow, and cone voltage were chosen in order to achieve maximum declustering without compromising spectral intensity.
We have demonstrated the ability of a single method on the UPLC/MS system to perform routine analysis and quality control of small interfering RNA molecules. Superior UPLC resolution, together with the mass accuracy capability of the Q-Tof Premier Mass Spectrometer, allows for assignment of the low-intensity peaks and deciphering of the RNAi oligonucleotide sequence.
This method’s exceptional dynamic range will enable the low-level quantitation of modifications, process impurities, or contaminants in the presence of the main RNAi molecule facilitating regulatory requirements compliance. Additionally, the comprehensive characterization and analysis results are generated by a single laboratory analyst on a single system, enabling greater overall laboratory throughput and efficiency.
720002412, January 2008