This is an Application Brief and does not contain a detailed Experimental section.
This application brief determines the collision cross-sections of singly charged lipid ions analyzed by MALDI mass spectrometry.
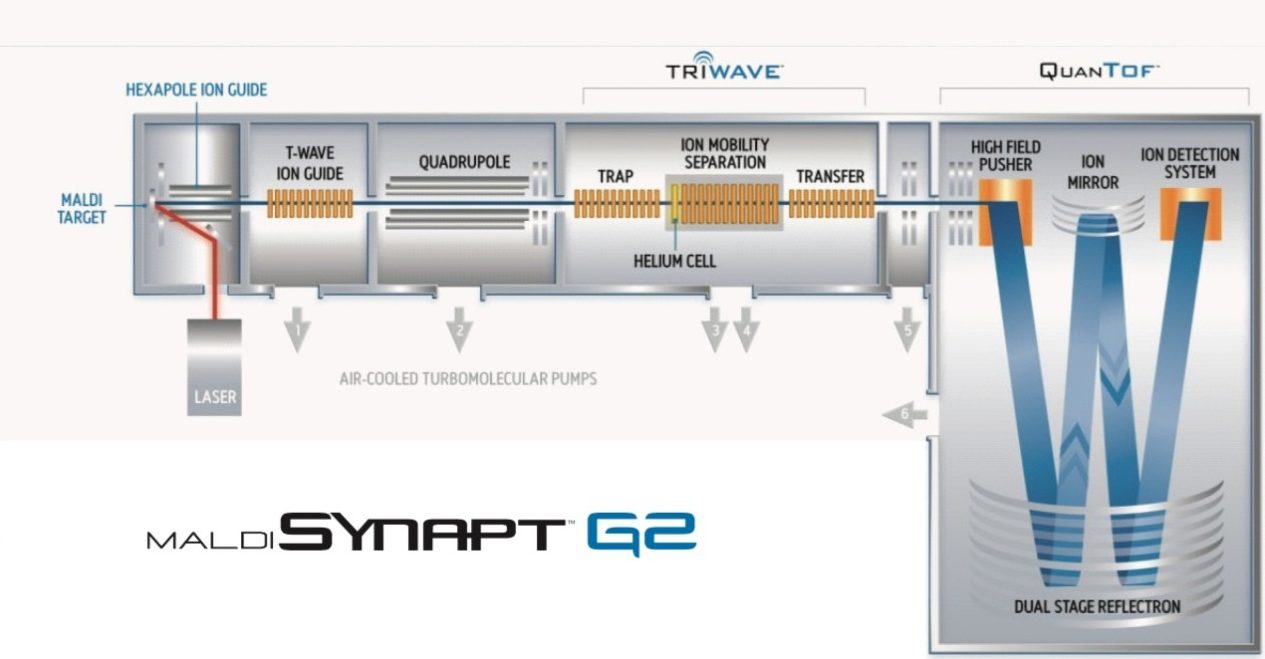
The MALD I SYNA PT G2 HDMS System provides an ideal platform to conduct lipid imaging studies, and in addition, allows the collision cross-sections to be calculated during the same experiment.
Lipidomics is a rapidly expanding field of research, where mass spectrometry plays a key role. Moreover, visualizing the localization of lipids within a tissue section is challenging since there are no antibodies specific to lipids. However, imaging by MALDI mass spectrometry allows the location of different classes of lipids directly from tissue sections to be visualized, thereby enhanc-ing lipid studies.
The use of ion mobility to evaluate the size and shape of ions in the gas phase is a technique which is rapidly gaining recognition. Initial studies were carried out on proteins; however, it has now been demonstrated that it is possible to use ion mobility to measure the collision cross-section of other types of molecules, like lipids.
The MALDI SYNAPT G2 HDMS System (schematic shown in Figure 1) provides an ideal platform to conduct these imaging studies, and in addition, allows the collision cross-sections to be calculated during the same experiment. Furthermore, data is acquired at high resolution, enabling exact mass measurements to be made on both precursor and fragment ions, coupled to the ability to separate target analytes from iso-baric background interferences using gas-phase ion mobility separations.
First, we calibrated the IMS T-Wave1 using polyalanine, with previously determined collision cross-section values (from http://www.indiana.edu/~clemmer). The calibration curve was generated with DriftScope Software using a power trend line where R=0.9995.
To calculate the collision cross-sections of each lipid standard, PC (16:0/16:0), PS (18:1/18:1), and PG (16:0/18:1) (Avanti Polar Lipids ) were mixed individu-ally with α-cyano-4-hydroxycinnamic acid (CHCA) matrix, spotted onto a MALDI stainless steel target, and analyzed under identical conditions to those used for the polyalanine standard. The analysis was performed using a 1 kHz Nd:YAG laser system on a MALDI SYNAPT G2 operated in HDMS mode. CCS calculations were also determined for the sodiated and potassiated species, when present.
Figure 2 shows that the collision cross-section, which was calculated to be 214.9 Å2 for MH+ of PC (16:0/16:0). This is in accordance with previously published results2 by Ridenour et. al., where their CCS calculation of these lipids on a MALDI SYNAPT (G1) HDMS instrument were found to be 215.3 Å2 +/- 3.6.
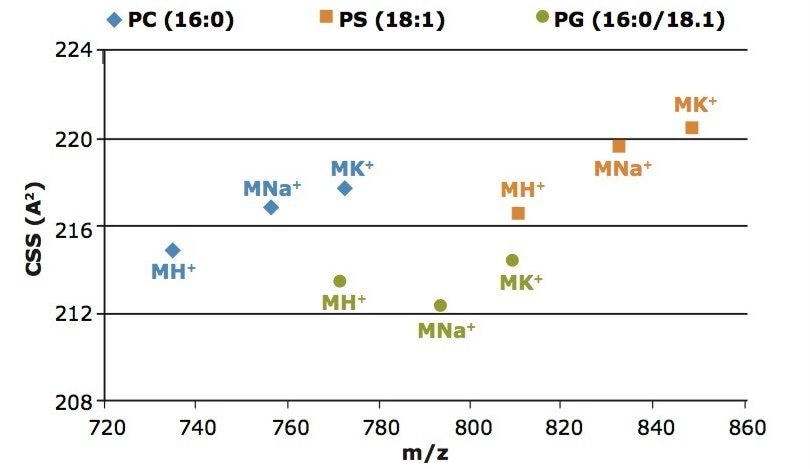
The collision cross-section of the Na+ and K+ species of the PC and PS standards indicate a more open configu-ration in the gas phase relative to the MH+ species when compared to the PG lipid standard. Here the Na+ and K+ of the PG ion appear to be more compact in the gas phase versus their MH+, which could indicate that the lipid may be folded around the salt.
Further analysis was carried out on a rat tissue section, under the same IMS conditions and the CCS of lipids calculated directly from tissue section, as shown in Figure 3. As lipids tend to have similar mass defects for each class, the data was colored following the first decimal of the m/z. Here, the trend lines can be observed in the data following the mass defect.
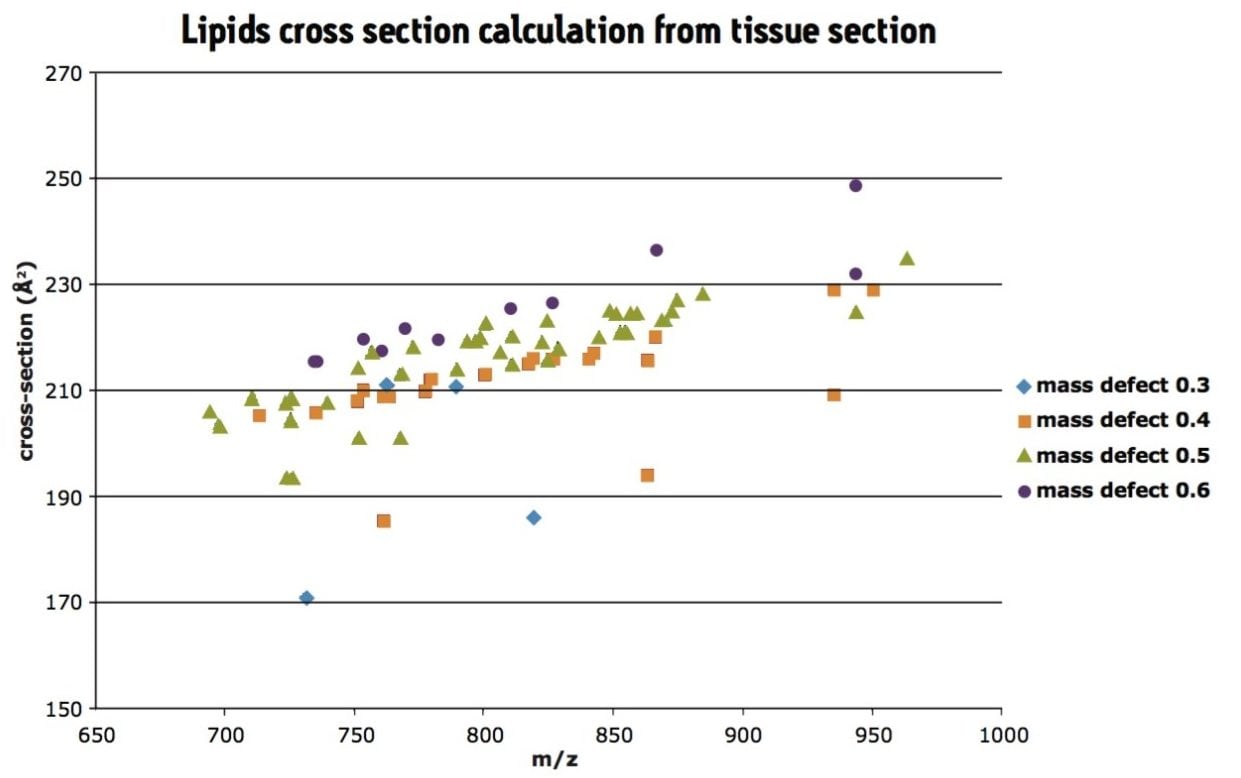
The MALDI SYNAPT G2 HDMS System enables the calculation of collision cross-sections of lipid standards. Here we have observed that the sodiated PG lipid standard had a more compact configuration in the gas phase compared to its counterpart MH+.
This type of analysis can also be carried out for endogenous species analyzed directly from tissue sections. In a single experiment, it is possible to calculate collision cross-sections for endogenous species and carry out a MALDI imaging experiment to observe their location throughout the tissue.
720003536, May 2010