Streamlined Characterization of Recombinant AAVs via Charge Detection Mass Spectrometry: Insights from the USP AAV8 Reference Standards
Rebecca J. D’Esposito, Anisha Haris, David Eatough, Vicki Knox, Ying Qing Yu, David Ballantyne, Stephen McDonald
Waters Corporation, United States
Published on October 13, 2025
Abstract
Recombinant adeno-associated viruses (rAAVs) are essential delivery vehicles for gene therapy. Comprehensive characterization of rAAV products is critical to ensure their safety and efficacy. However, the intrinsic heterogeneity of rAAVs presents significant analytical challenges. Charge Detection Mass Spectrometry (CDMS) addresses these challenges by directly measuring the mass of individual ions through their charge and mass-to-charge ratio (m/z). This measurement enables high-resolution characterization of intact rAAV particles. In this application note, the use of a commercial electrostatic linear ion trap (ELIT) CDMS is demonstrated to characterize the USP rAAV8 empty and full reference standards by determining particle mass, calculating the empty/full ratio, and identifying impurities such as partially filled particles. Furthermore, it showcases how the dedicated CDMS waters_connect™ software, CDMS Toolkit, streamlines the analysis workflow for both GxP and non-GxP laboratory environments, facilitating efficient calculation of both the empty/full ratio and impurity levels within AAV samples.
Benefits
- Xevo™ CDMS enables high-resolution mass analysis of intact rAAV particles, quantifying empty, partial, and full capsids
- Integrated with waters_connect, it streamlines reporting and boosts throughput
- Its sensitivity enhances impurity detection, improving quality control and accelerating gene therapy development for clinical and commercial readiness
Introduction
rAAVs are among the most widely used vectors for gene therapy, owing to their low immunogenicity, broad tissue tropism, and ability to mediate long-term gene expression.1 As rAAV therapeutics progress toward clinical and commercial development, robust analytical techniques must address their structural diversity and compositional heterogeneity.1 Conventional mass spectrometry methods often fall short when analyzing large, heterogeneous viral particles like rAAVs, as charge deconvolution isn’t possible due to the low resolution of these analytes in the m/z region. In contrast, CDMS is a powerful technique that allows direct measurement of both the mass and charge of individual ions, eliminating the need for high m/z resolution to analyze these species.2 CDMS offers a transformative approach to rAAV characterization by providing high-resolution, single-particle mass measurements without the need for extensive sample preparation or separation steps.2 By resolving heterogeneous populations within a single sample, CDMS offers insights into genome packaging efficiency, capsid integrity, and the presence of partially filled or aggregated rAAV species.2
A major advance in this field is the availability of a commercial CDMS system: the Xevo CDMS, which is equipped with dedicated, robust software on the waters_connect platform. CDMS Toolkit, a waters_connect application, performs data acquisition and automated (Figure 1) analysis of data acquired by Xevo CDMS. This integration enables streamlined, high-resolution analyses of intact rAAV particles, while also facilitating routine operation in laboratory settings.
This application note outlines the core principles of CDMS and demonstrates its effectiveness in the comprehensive analysis of rAAV-based therapeutics. Using commercial rAAV8 reference standards from USP as a case study, the advantages of CDMS is highlighted over conventional methods and its potential to streamline quality control in gene therapy development.
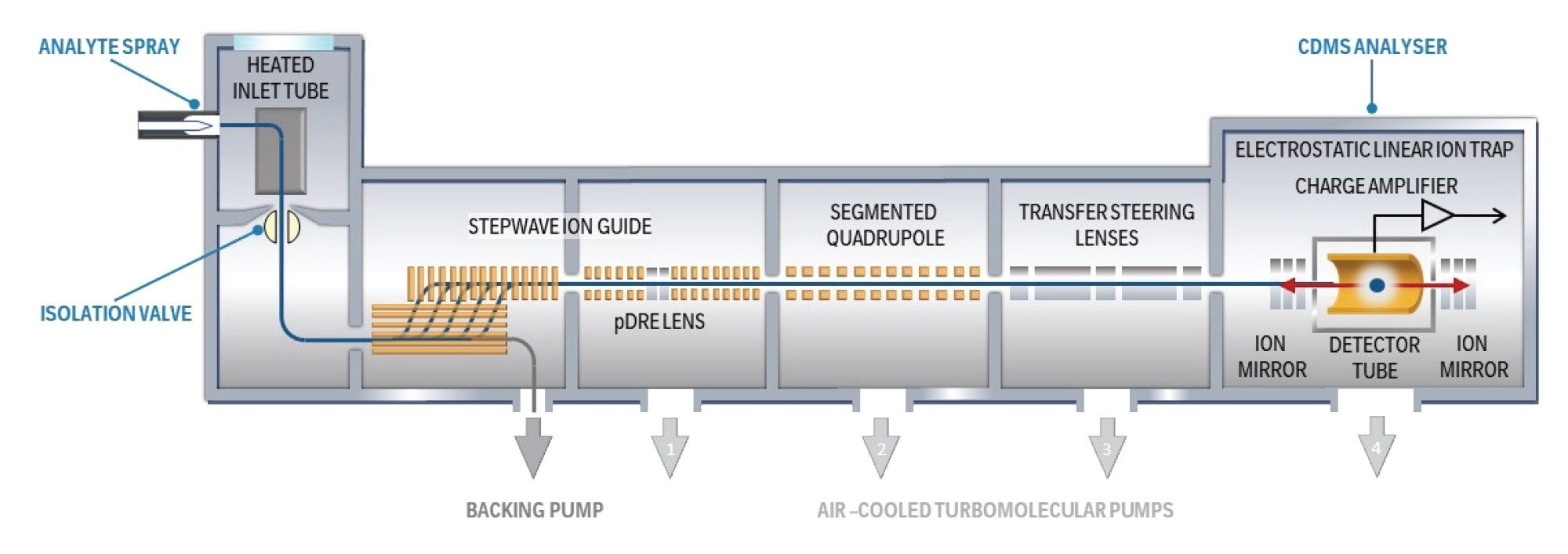
Experimental
Sample Preparation
AAV8 empty and full capsids were purchased from USP [cat. 1000301(empty) and 1000302 (full)]. Orthogonal data was sourced from the standards’ certificates.6-7 The samples were buffer exchanged into 200 mM ammonium acetate solution with 0.01 % Pluronic™ F-68 (Gibco) using BioSpin® P-6 size-exclusion columns (Bio-Rad Laboratories, Inc), following manufacturer’s protocol. Sample preparation for each standard was performed in triplicate, once per aliquot.
All data were acquired within the waters_connect informatics platform (version 4.2.0) using the CDMS Toolkit application and processed within the same application.
MS Conditions
|
MS system: |
Xevo Charge-Detection Mass Spectrometry System |
|
|
Acquisition mode: |
Full Scan |
|
|
Ionization mode: |
Positive Ion |
|
|
Capillary voltage: |
2.0–2.2kV |
|
|
Cone voltage: |
50 |
|
|
Trapping time: |
100 ms |
|
|
Acquisition duration: |
15–20 minutes |
|
|
MS software: |
waters_connect software ver. 4.2.0 |
Data Processing Workflow
All data presented here was processed using CDMS Toolkit, a waters_connect application. CDMS Toolkit is organized into three main sections: Investigate Data, Methods, and Analysis.
The Investigate Data section allows users to view acquired CDMS data. For each injection, CDMS displays the charge, m/z, and mass information for each ion as histograms. These three domains of data can be compared using two-dimensional plots for deeper investigation (see Figure 2). By leveraging all three domains, researchers gain a more comprehensive understanding of complex particle distributions, and the components present in their samples.
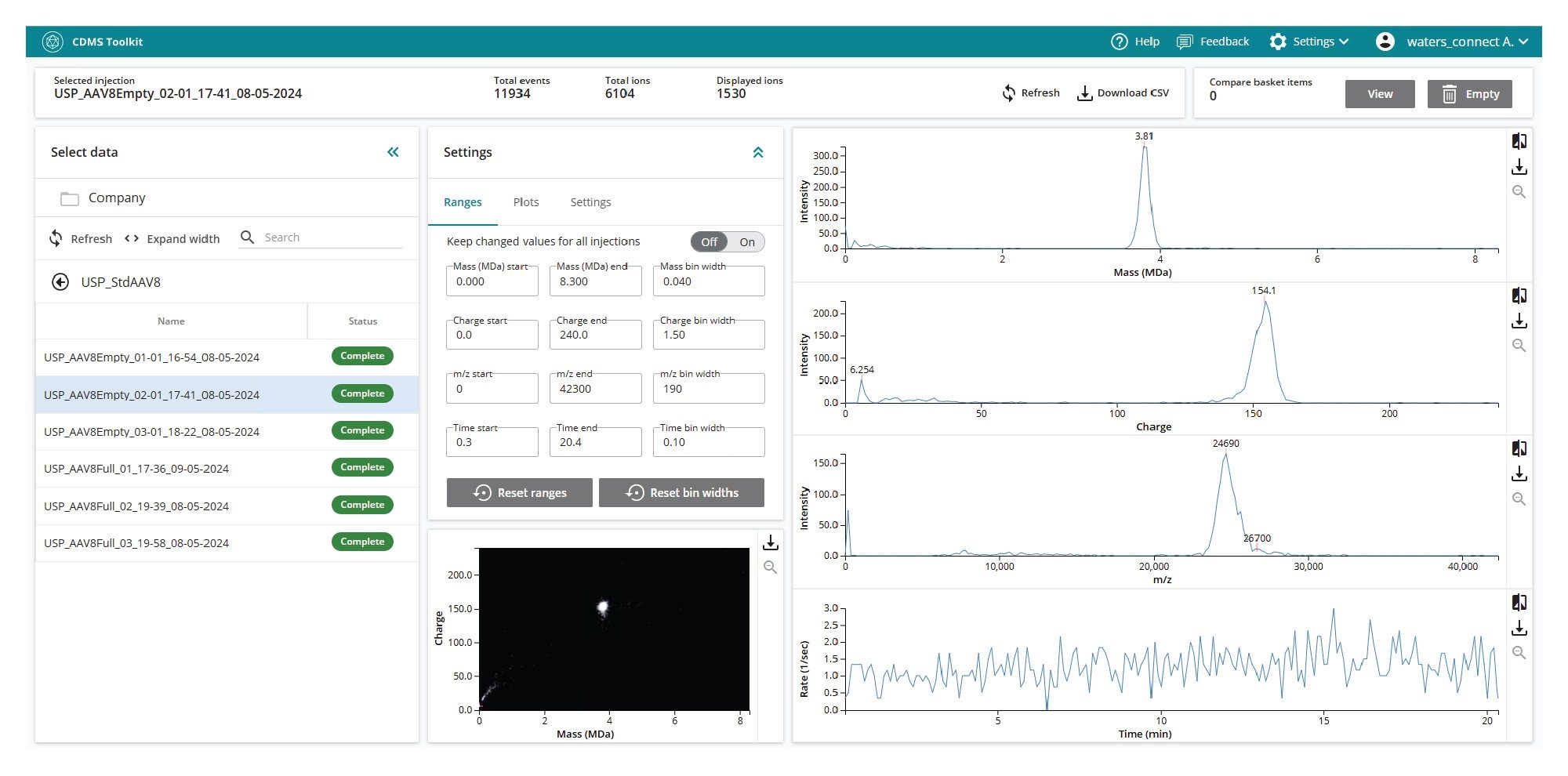
CDMS Toolkit is a flexible and powerful application capable of supporting a wide range of scientific workflows. One key implementation is the determination of rAAV capsid content ratios, which is a critical quality attribute (CQA) essential for evaluating the therapeutic efficacy and minimizing potential safety risks for rAAV-based therapeutics.3 To determine capsid content ratios in an rAAV sample, a method must first be created and defined within the Methods section of the CDMS Toolkit (see Figure 3). To enable relative quantitation of rAAV species, such as empty, partial, and full capsids, specific ranges for mass, charge, and m/z must be defined for each species. For example, a full AAV8 particle may be characterized by the following boundaries: 5.0–5.2 MDa (mass), 140–160e (charge), and 23,000–35,000 m/z. Ions that simultaneously meet all three criteria are classified as “full AAV8” and contribute to the calculated full-capsid percentage. Once the method has been configured, it can be published for use in subsequent analyses, with access permissions managed at the user level to prevent unauthorized modifications or viewing.
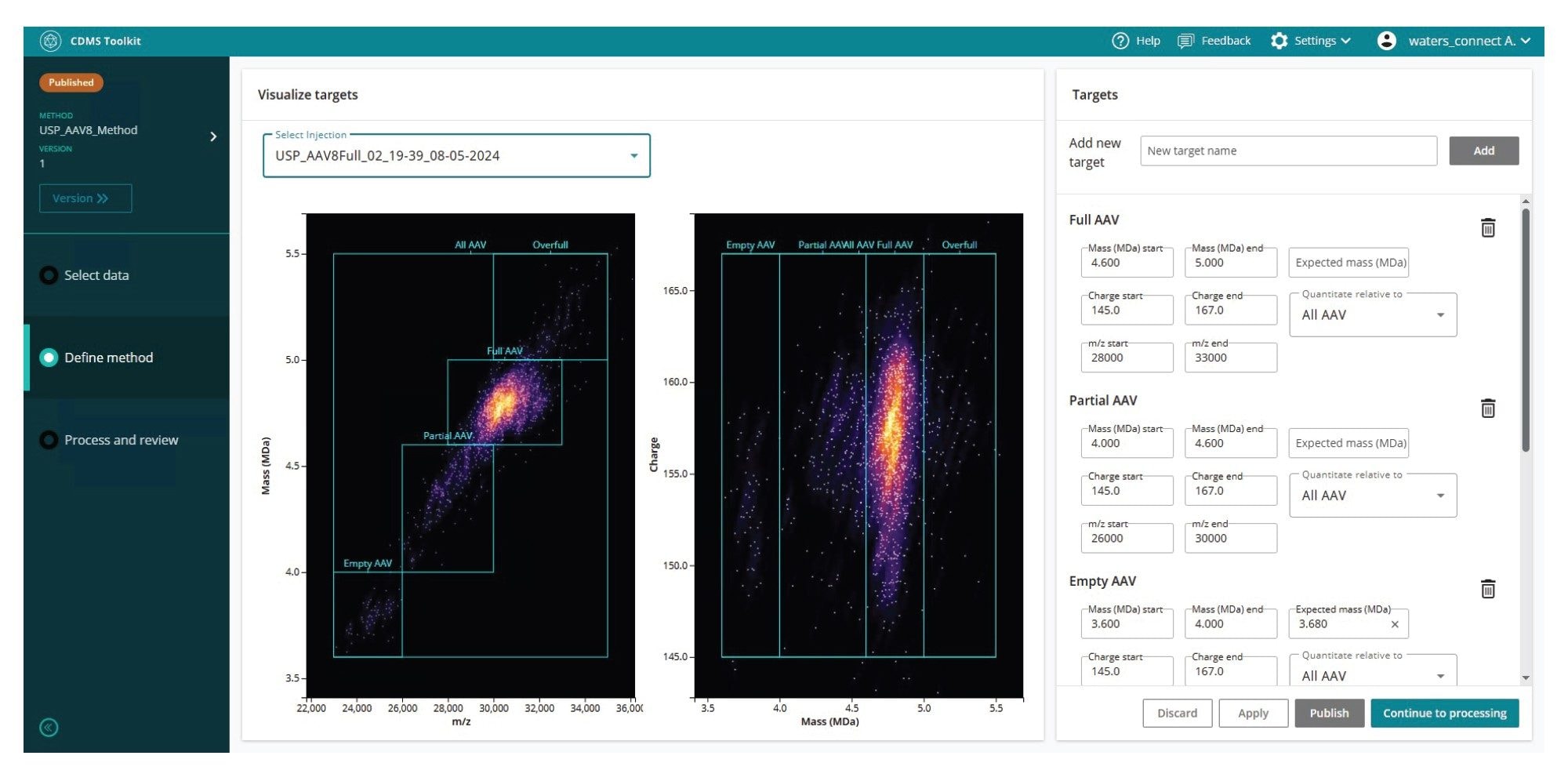
Using the method defined in the previous step, an Analysis can be performed over multiple injections. After selecting the data, applying the method, and processing the results, the output is displayed in a table format. This results table can be exported as a CSV file and imported into any preferred data processing software. Trends across injections are visualized as bar graphs for each species defined in the method. These graphs appear on the right-hand side of the interface, adjacent to the results table. Additionally, an analysis report is generated for each processed analysis which can be downloaded in PDF format. This report includes the results table, trend bar graphs, and histograms for the mass and charge domains for every species defined in the method across all acquisitions included in the analysis.
Results and Discussion
Determining the capsid content ratios of rAAV8 USP reference standards (empty and full) by CDMS
Determining the full-to-empty capsid ratio is a CQA for rAAV therapeutics, directly impacting potency, safety, and dosing consistency.3 Traditional methods such as sedimentation velocity analytical ultracentrifugation (sv-AUC), mass photometry (MP), and size-exclusion chromatography with multi-angle light scattering (SEC-MALS) provide useful information but are often limited in resolution, throughput, or accuracy.3 CDMS overcomes these challenges by simultaneously measuring both the m/z and absolute charge of individual ions, allowing direct determination of particle mass.2 This enables unambiguous separation of empty and full capsids based on their distinct mass difference. With minimal sample preparation, high sensitivity, and robust quantification in complex mixtures, CDMS provides a precise and efficient approach for capsid content analysis, uniquely suited to the rapid analytical needs of gene therapy development.
Two rAAV8 reference standards, empty and full, were characterized using CDMS. These standards, purchased from USP, were accompanied by documentation containing capsid content ratios and supporting data from orthogonal techniques, including sv-AUC, MP, and SEC-MALS. Prior to CDMS analysis, both standards were buffer-exchanged into 200 mM ammonium acetate containing 0.01% Pluronic F-68. Samples were introduced into the CDMS instrument via nano-electrospray ionization (nESI) in positive ion mode, using 5 µm borosilicate glass emitters. A capillary voltage between 2.0 and 2.2 kV was applied. The CDMS trap detector was configured to trap ions every 100 ms, and data acquisition was performed over a period of 15 to 20 minutes.
Figure 4A shows a CDMS mass histogram for the empty rAAV8 capsid standard. A dominant peak appears at 3.81 MDa with a FWHM (full width half maximum) of 0.16 MDa, consistent with the documented theoretical mass of ~3.8 MDa (0.6% deviation). The average charge observed for empty capsids is 154e (Figure 4B). Additional species are detected at ~2 MDa and between 3–6 MDa. Signals below 2 MDa are likely process-related impurities, while those in the 3–6 MDa range share the same charge population as capsids, suggesting similar ionization and structure. These species represent ≤1.5% of ion counts in the rAAV mass region and are attributed to partially filled or full capsids. Using the analysis workflow as described in the experimental section, the capsid content ratios for this preparation were calculated as 98.5% empty, 0.9% partially filled, and 0.6% full capsids.
Figure 4C shows the CDMS mass histogram for the full rAAV8 capsid standard. The dominant peak at 4.79 MDa (FWHM 0.20 MDa) is attributed to full capsids, consistent with the reported theoretical mass of ~4.6 MDa, with a 2.67% mass deviation. A secondary peak at 3.84 MDa (FWHM 0.17 MDa) corresponds to empty capsids, with ~1% deviation. Both peaks exhibit an average charge of 156e (Figure 4D). Additional partially resolved features between 4.0–4.6 MDa and 5.0–5.5 MDa also carry ~155e charge and are assigned to partially filled and overfull capsids. These indicate product-related impurities. Based on the software and analysis described, the capsid distribution in the “full” rAAV8 standard was 4% empty, 16% partially filled, 77% full, and 3% overfull.
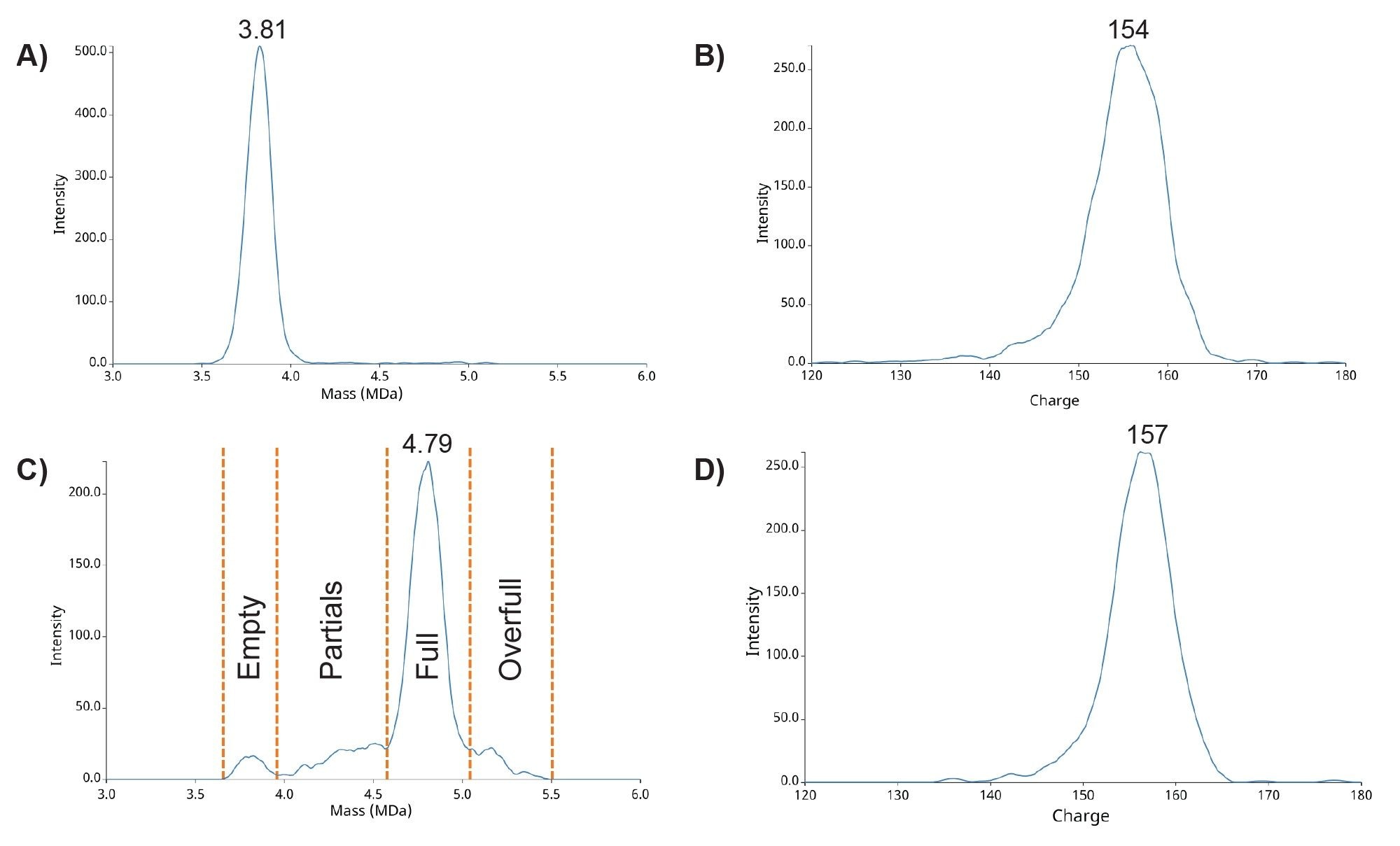
Table 1B compares the rAAV8 capsid ratios for the full capsid standard determined by multiple techniques, including CDMS. While results for the empty capsid standard were consistent across all analytical techniques (Table 1A), notable discrepancies were observed for the full capsid standard, even within the supplier’s own documentation.6-7 These inconsistencies stem from differences in analytical principles: MP and CDMS measure individual particles, whereas SEC‑MALS and sv‑AUC assess bulk solution properties.4 Both sv‑AUC and CDMS can resolve partially filled and other capsid species, a capability that SEC‑MALS and MP generally lack. Consequently, CDMS and sv‑AUC often agree on detailed capsid distributions, though sv‑AUC can under‑ or over‑resolve partial species depending on heterogeneity, curve fitting, and genomic size.5 The divergence observed between CDMS and sv-AUC is unusual but may be attributed to the smaller-than-typical genome size used in these reference standards (2.8 kb compared to the typical 4.7–5.0 kb). Ultimately, when partially filled, full, and overfull species are grouped together, the reported full‑capsid content align across methods: 96% by CDMS, 97% by sv‑AUC, 98% by SEC‑MALS, and 91% by MP.
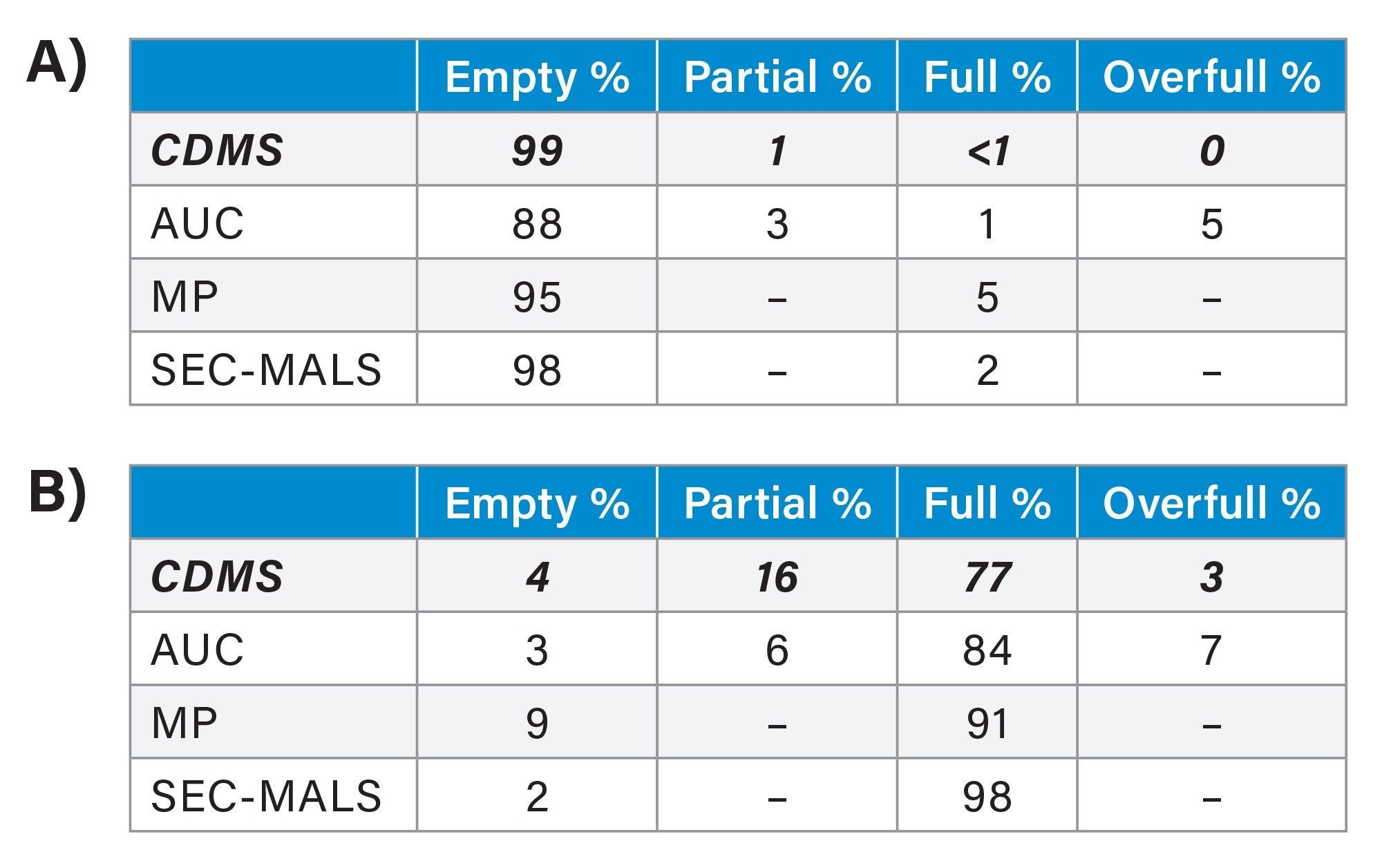
Table 1. A) Capsid content ratios for empty AAV8 standard and B) full AAV8 standard. By row 1: CDMS, row 2: AUC, row 3: MP, and row 4: SEC-MALS.
- Indicates not measured
Conclusion
The Xevo CDMS is an advanced commercial analytical platform designed for the direct and automated mass measurement and characterization of large, complex biomolecules. With the dedicated CDMS Toolkit application, a streamlined analytical workflow was developed for analyzing rAAV capsid ratios. CDMS data is highly informative, offering multiple dimensions of data that provide a comprehensive view of each injection. Although CDMS Toolkit was used for rAAV capsid analysis in this study, its capabilities extend beyond analysis of rAAVs. The method’s targets can be broadly defined to accommodate any analyte of interest.
The USP rAAV8 reference standards case study highlights that rAAV capsid characterization can vary across analytical techniques in their ability to distinguish between empty, partial, and full capsids. Agreement across analytical techniques improves when partial and overfull capsids are grouped with full capsids for direct comparison. For objective assessment, it is essential to understand the resolution limits of each analytical technique to reconcile differences in capsid ratio measurements. sv-AUC provides high-resolution separation based on sedimentation properties, MP offers rapid single-particle analysis with moderate sensitivity, SEC-MALS excels in molecular weight determination but struggles to resolve closely related capsid species, while CDMS uniquely combines charge and mass measurements to deliver high-resolution, particle-level characterization of heterogeneous populations.
References
- Pupo A, Fernández A, Low SH, François A, Suárez-Amarán L, Samulski RJ. Aav vectors: The Rubik’s cube of human gene therapy. Molecular Therapy. 2022 Dec;30(12):3515–41. doi:10.1016/j.ymthe.2022.09.015.
- Jarrold MF. Applications of charge detection mass spectrometry in Molecular Biology and Biotechnology. Chemical Reviews. 2021 Oct 12;122(8):7415–41. doi:10.1021/acs.chemrev.1c00377.
- Kontogiannis T, Braybrook J, McElroy C, Foy C, Whale AS, Quaglia M, et al. Characterization of aav vectors: A review of analytical techniques and critical quality attributes. Molecular Therapy - Methods & Clinical Development. 2024 Sept;32(3):101309. doi:10.1016/j.omtm.2024.101309.
- Werle AK, Powers TW, Zobel JF, Wappelhorst CN, Jarrold MF, Lyktey NA, et al. Comparison of analytical techniques to quantitate the capsid content of adeno-associated viral vectors. Molecular Therapy - Methods & Clinical Development. 2021 Dec;23:254–62. doi:10.1016/j.omtm.2021.08.009.
- Hayes DB, Dobnik D. Commentary: Multiplex dPCR and SV-AUC are promising assays to robustly monitor the critical quality attribute of AAV Drug Product Integrity. Journal of Pharmaceutical Sciences. 2022. Aug;111(8):2143–8. doi:10.1016/j.xphs.2022.04.010.
- [AAV8 (empty capsids) (300 ul) (International Cold chain shipment required)] - cas [N/A] [Internet]. [cited 2025 Sept 2]. Available from: https://store.usp.org/product/1000301.
- [AAV8 (full capsids) (300 UL) (International Cold chain shipment required)] - cas [N/A] [Internet]. [cited 2025 Sept 2]. Available from: https://store.usp.org/product/1000302.
Featured Products
720009054, September 2025