Advancing ADC Characterization: SEC-Based Native DAR and Drug Distribution Analysis Using Multi-Reflecting TOF-MS and INTACT Mass Application
Jonathan Fox, Scott J Berger, Laetitia Denbigh, Sam Ippoliti
Waters Corporation, United States
Published on December 24, 2025
Abstract
This technology brief describes the application of the ACQUITY™ Premier UPLC™ System coupled with the Xevo™ MRT Mass Spectrometer for the native mass analysis of monoclonal antibodies (mAbs) and antibody-drug conjugates (ADCs). Leveraging the INTACT Mass Application within the waters_connect™ platform, the workflow enables fully integrated data acquisition, automated mass deconvolution, and streamlined review and reporting. Using an FDA approved cysteine-conjugated ADC as a model system, the study demonstrates the capabilities of the Xevo MRT Mass Spectrometer and INTACT Mass 1.9 Application for concurrent glycoform profiling and drug-to-antibody ratio (DAR) analysis, supporting confident identification and quantification of product variants. Together, this integrated solution provides a robust and efficient approach for comprehensive biotherapeutic characterization.
Benefits
- Automated workflow for a complex analysis
Automatically calculate DAR and drug distribution with the INTACT Mass Application version 1.9. This approach streamlines mAb conjugate analysis by reducing manual efforts, minimizing interpretation errors, and making advanced ADC workflows accessible and consistent for analysts with varying levels of LC-MS experience. The result is faster productivity, greater consistency, and a reduced training burden.
- High-quality data for confident decisions
Gain detailed insights into glycoform profiles, aggregates, and DAR values, all in a single LC-MS run, using high-resolution native SEC-MS with the Xevo MRT Mass Spectrometer. This powerful setup delivers the sensitivity and accuracy needed for comprehensive product understanding, enabling faster decisions and confident CQA assessments.
Introduction
ADCs are a rapidly expanding class of targeted biotherapeutics, combining the specificity of mAbs with the cytotoxic potency of small-molecule drugs. A critical quality attribute of ADCs is the DAR, which directly influences therapeutic efficacy, pharmacokinetics, and safety. Traditional methods for DAR determination, such as UV spectroscopy or hydrophobic interaction chromatography (HIC), often lack the resolution needed to fully characterize the complexity and heterogeneity of ADCs.
Native mass spectrometry (MS) when coupled with size exclusion chromatography (SEC), provides a powerful, label-free approach for ADC analysis. SEC preserves native like conditions while separating product size variants, and combined with high-resolution native MS analysis, enables mass confirmation of glycoform profiles and drug-conjugate antibody variants. This combined approach supports direct quantification of glycoform ratios, detection aggregates, and degradation products, aa well as the assessment of DAR value and drug distribution, all within a single streamlined LC-MS workflow.
The integration of multi-reflecting time-of-flight MS (Figure 1) into SEC-MS workflows further enhances analytical performance, delivering high-quality data with excellent sensitivity, resolution, and mass accuracy. When paired with the waters_connect platform and the INTACT Mass App, this setup supports automated data acquisition, data processing, and reporting.
The INTACT Mass App, within the compliance-ready waters_connect ecosystem, for intact proteins,1 oligonucleotides,2–3 and synthetic peptides.4 Versions 1.9 and later includes advanced features needed for ADC analysis, such as customizable deconvolution settings and integrated DAR calculations. These capabilities are fully embedded and automated, reducing complexity, minimizing errors, and enabling confident ADC analysis on a compliance-ready platform.
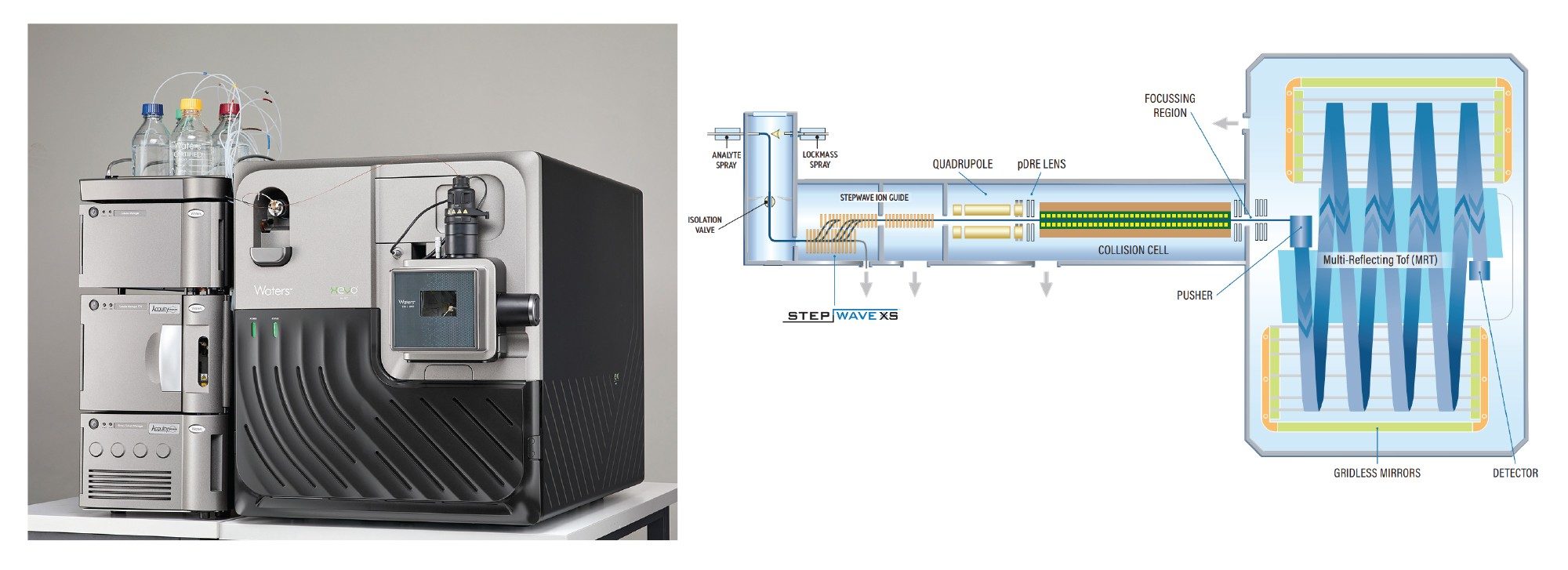
Experimental
Sample Preparation
Herceptin™ (trastuzumab) (unconjugated fam-trastuzumab-deruxtecan-nxki) and the cystine-conjugated ADC, ENHURTU™ (fam-trastuzumab-deruxtecan-nxki), 20 µg/ml in vial, were prepared by adding 90 µl Milli-Q water to a 10 µl sample, resulting in a concentration of 2 µg/µl.
LC Conditions
|
LC system: |
Waters ACQUITY Premier UPLC System (Binary) |
|
|
Detection: |
UV 280 nm |
|
|
Vials: |
TruView LCMS Certified 12 x 32 mm Screw Neck Vial, Total Recovery, with Cap and Preslit PTFE/Silicone Septum (p/n: 186005663CV) |
|
|
Column: |
ACQUITY UPLC Protein BEH™ SEC Column ,200 Å, 1.7 µm 2.1 mm x 150 mm ( p/n: 186008471) |
|
|
Column temperature: |
30.0 °C |
|
|
Sample temperature: |
6.0 °C |
|
|
Injection volume: |
2 µL |
|
|
Flow rate: |
0.100 ml/min |
|
|
Mobile phase A: |
10 mM Ammonium acetate |
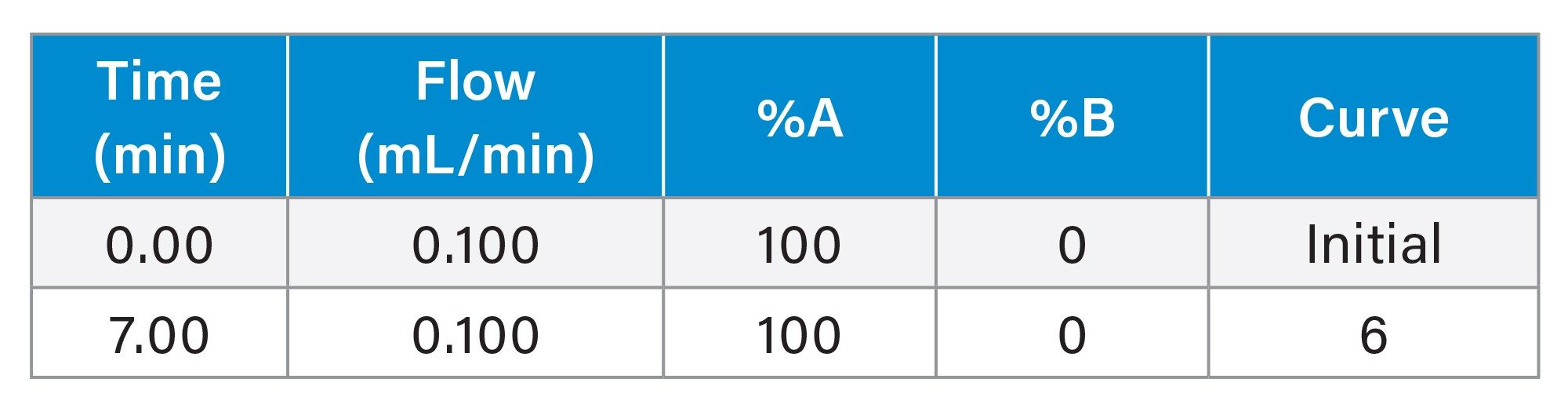
Gradient Table
MS Conditions
|
MS system: |
Waters Xevo MRT Mass Spectrometer |
|
Mode: |
MS |
|
Mass range: |
500-8000 m/z |
|
Polarity: |
Positive |
|
Scan rate |
1 Hz |
|
Cone voltage: |
120 V |
|
Cone gas: |
50 L/hr |
|
Source temperature: |
100 °C |
|
Desolvation temperature: |
450 °C |
|
Desolvation gas: |
1200 L/hr |
|
Capillary voltage: |
2.20 kV |
Results and Discussion
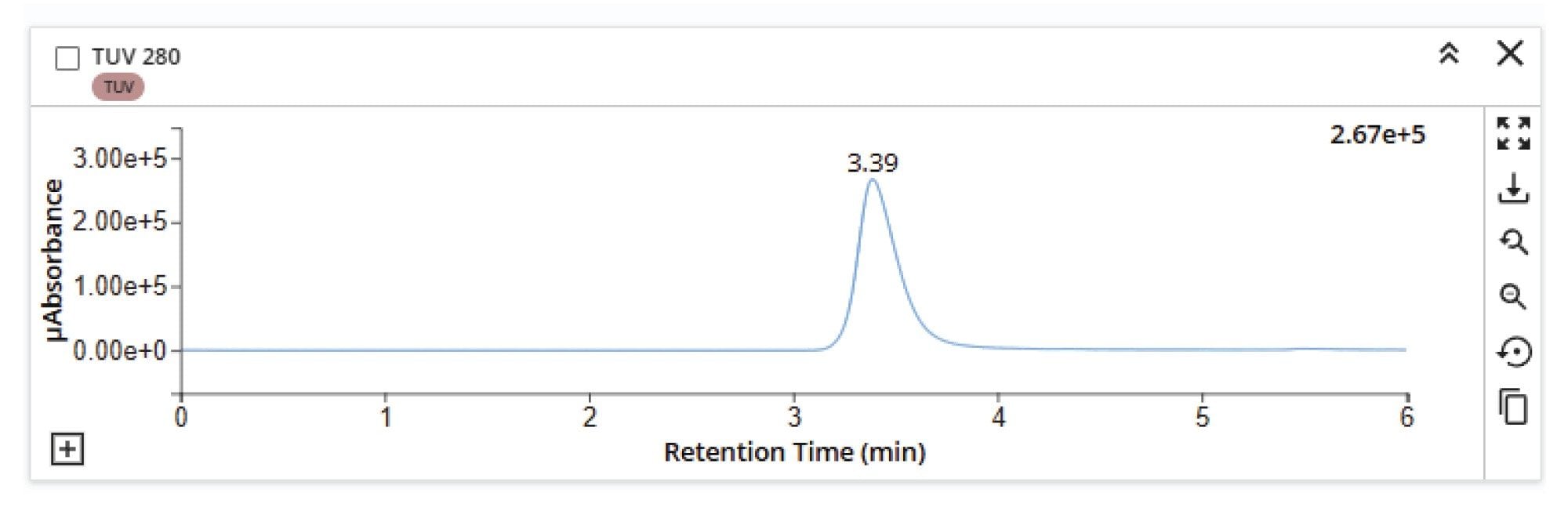
SEC-MS analysis of a Herceptin (trastuzumab) sample under native conditions demonstrates effective desalting of the main mAb peak and potential for resolution from any low- and high-molecular-weight size variant species (Figure 2). The size exclusion chromatogram shows a dominant monomeric peak, confirming the structural integrity of the sample. Mass spectral data under this peak reveals a well-resolved native charge envelope corresponding to the intact mAb (Figure 3), with deconvoluted spectra (Figure 4) indicating a simple distribution of biantennary Herceptin (trastuzumab) glycoforms.
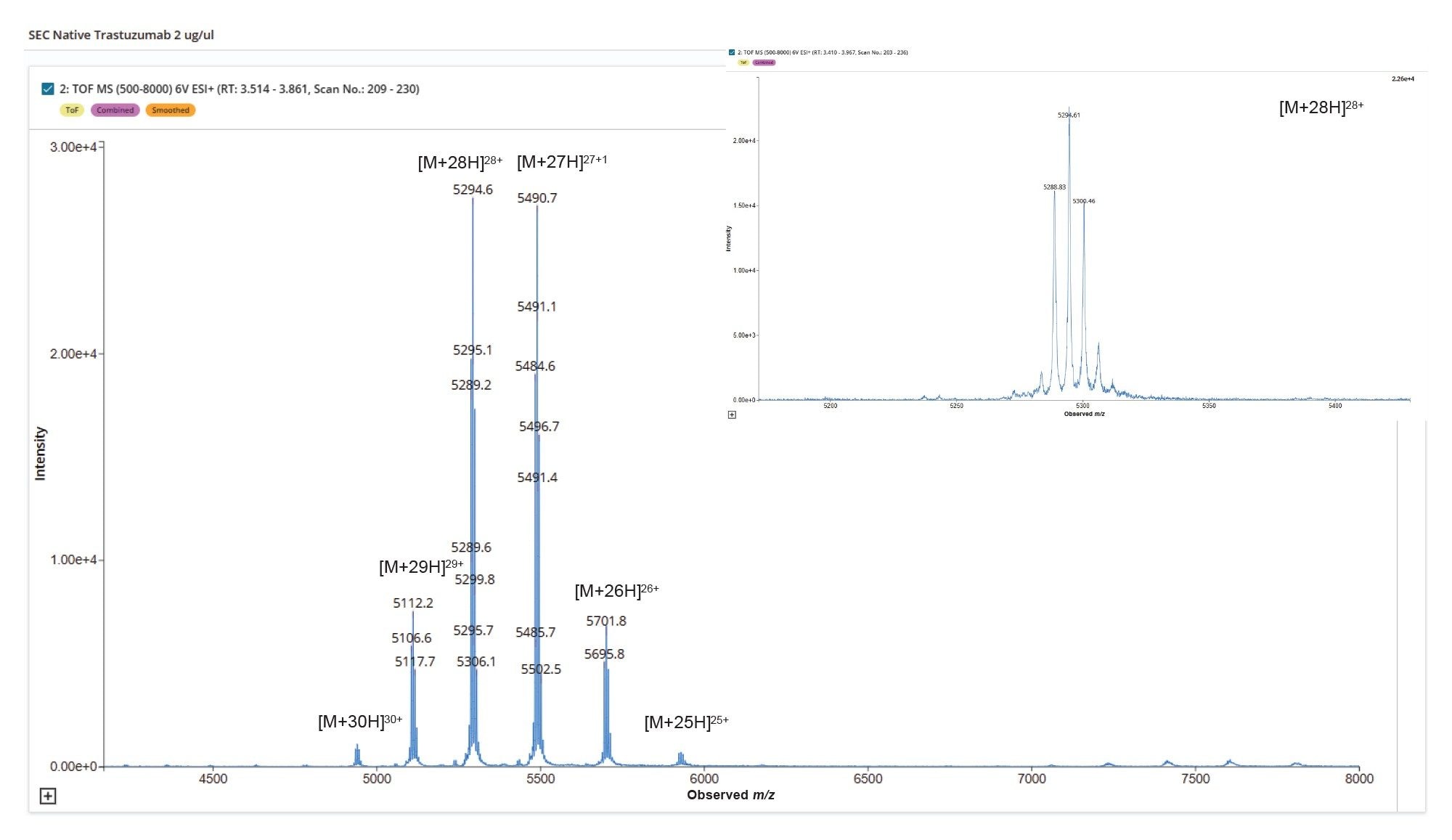
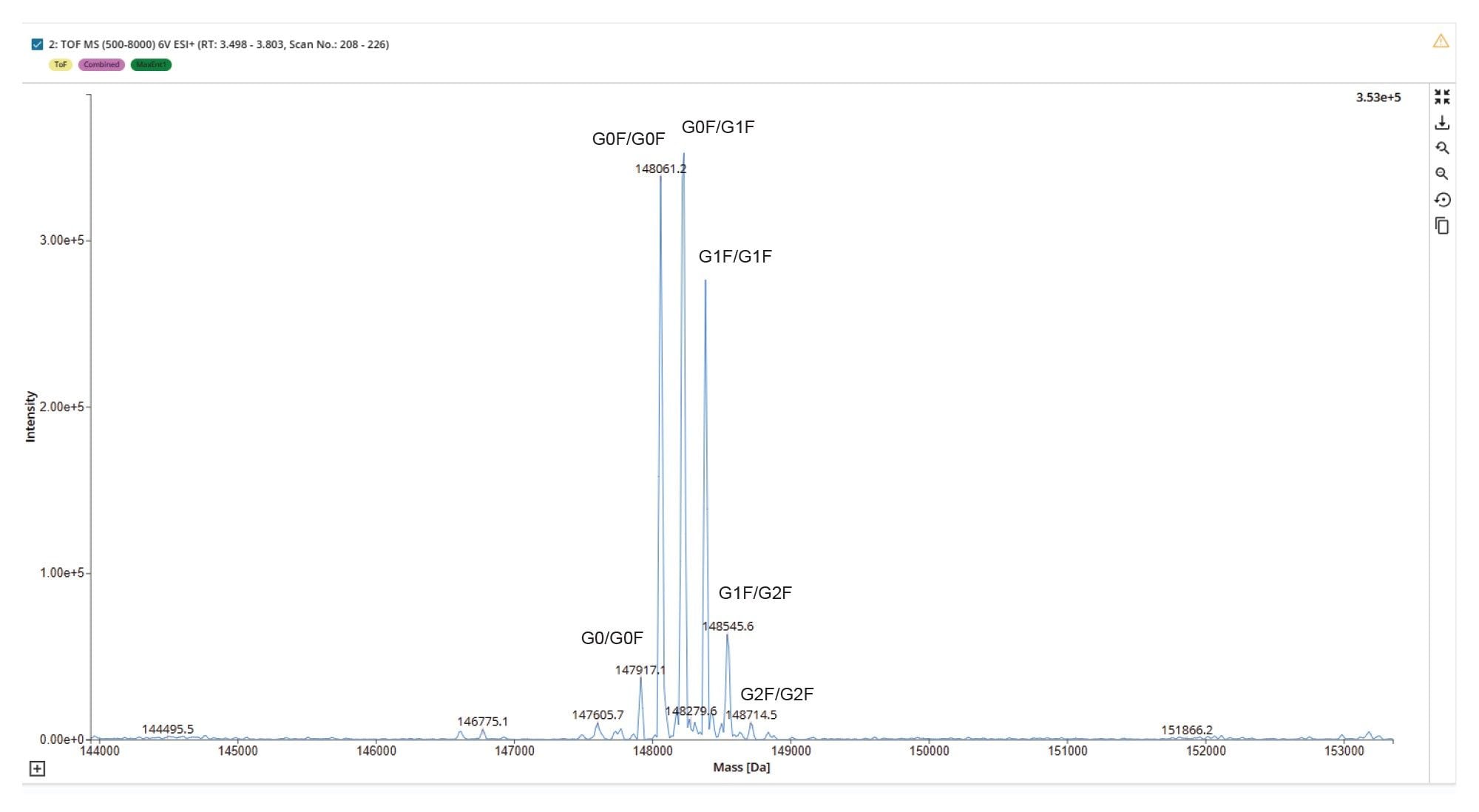
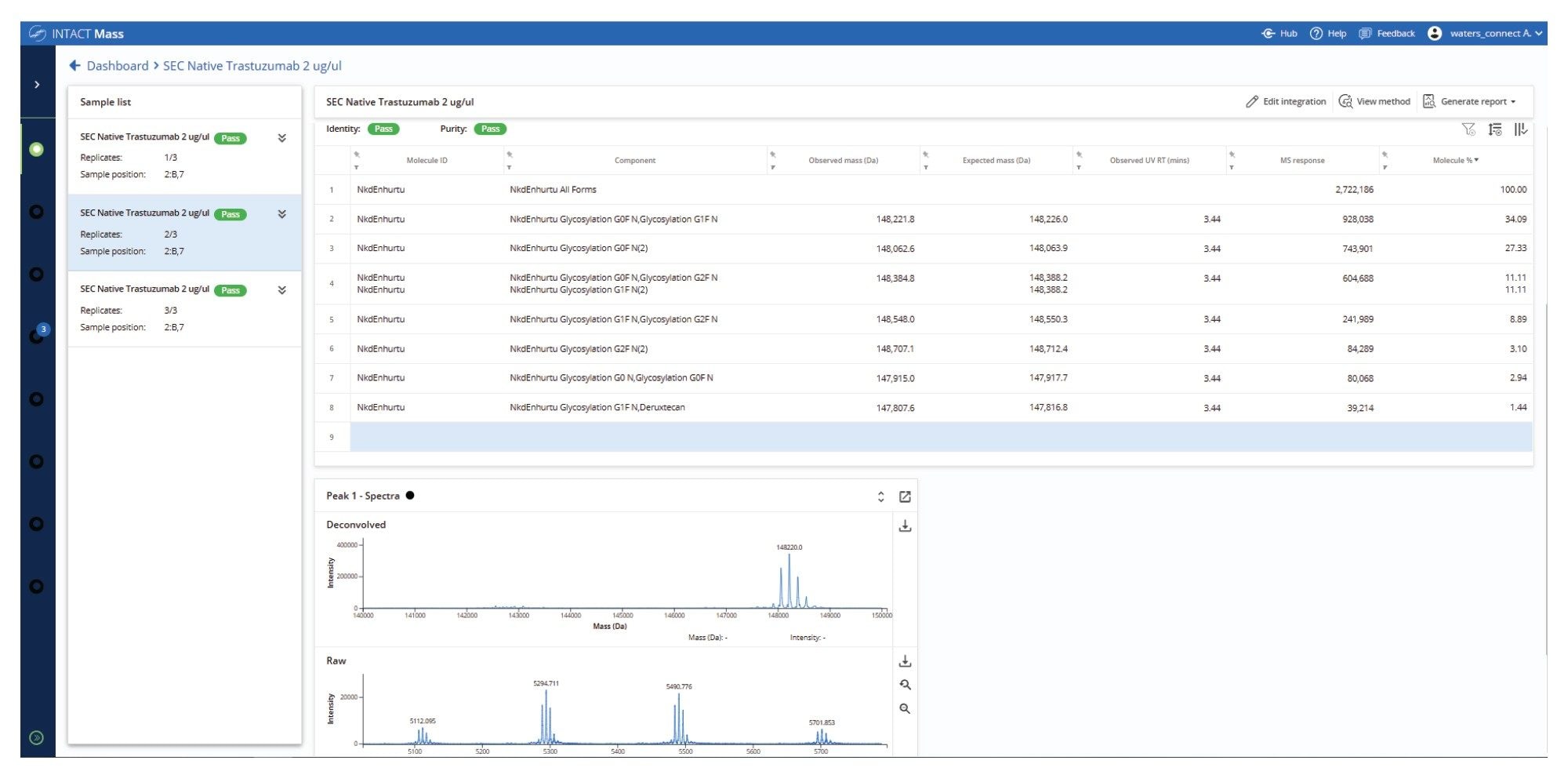
These results confirm the structural integrity of the sample and highlight the suitability of the SEC-Xevo MRT Mass Spectrometer workflow for preserving native biotherapeutic conformations. This enables consistent detection of higher ADC forms that may potentially interfere with Cys bonds connecting antibody subunits in the Cys ADC modified molecule. The high-quality spectral profile of Herceptin (trastuzumab) enables automated and accurate determination of N-glycoform profile utilizing the INTACT Mass Application (Figure 5) to automate peak detection and deconvolution processing and provides a robust analytical baseline for subsequent ADC DAR payload characterization.
Analysis of a Cystine-Conjugated ADC, ENHERTU™ (fam-trastuzumab-deruxtecan-nxki)
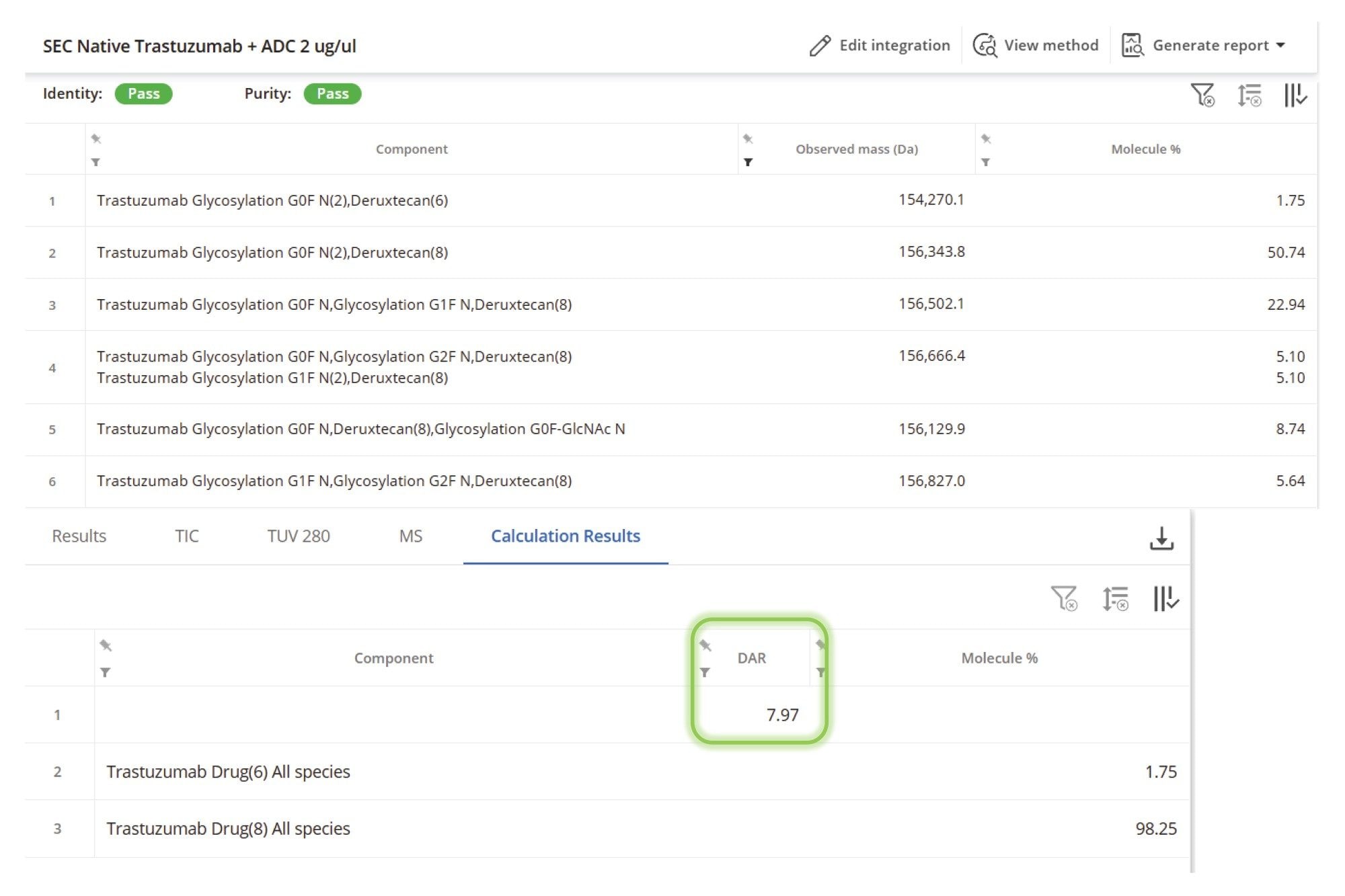
A critical quality attribute of ADCs and other protein conjugates is assessing the variability of drug payload molecules conjugated to the protein. This is reflected in both drug distribution (the relative percentage of each conjugated species) and overall DAR value (a single value derived from the weighted average of each conjugated species). The automation of these calculations in the INTACT Mass App version 1.9 enables consistent generation and direct access to these key quality measures. Within the processing method, the drug/payload modification is indicated with a specific tag, called “Drug”. During mass spectral data processing, the software automatically matches observed masses to theoretical masses of conjugated species. The relative MS responses for matched components with a “Drug” tag attached (along with the level of unconjugated protein) are then used for the DAR and drug distribution calculations.
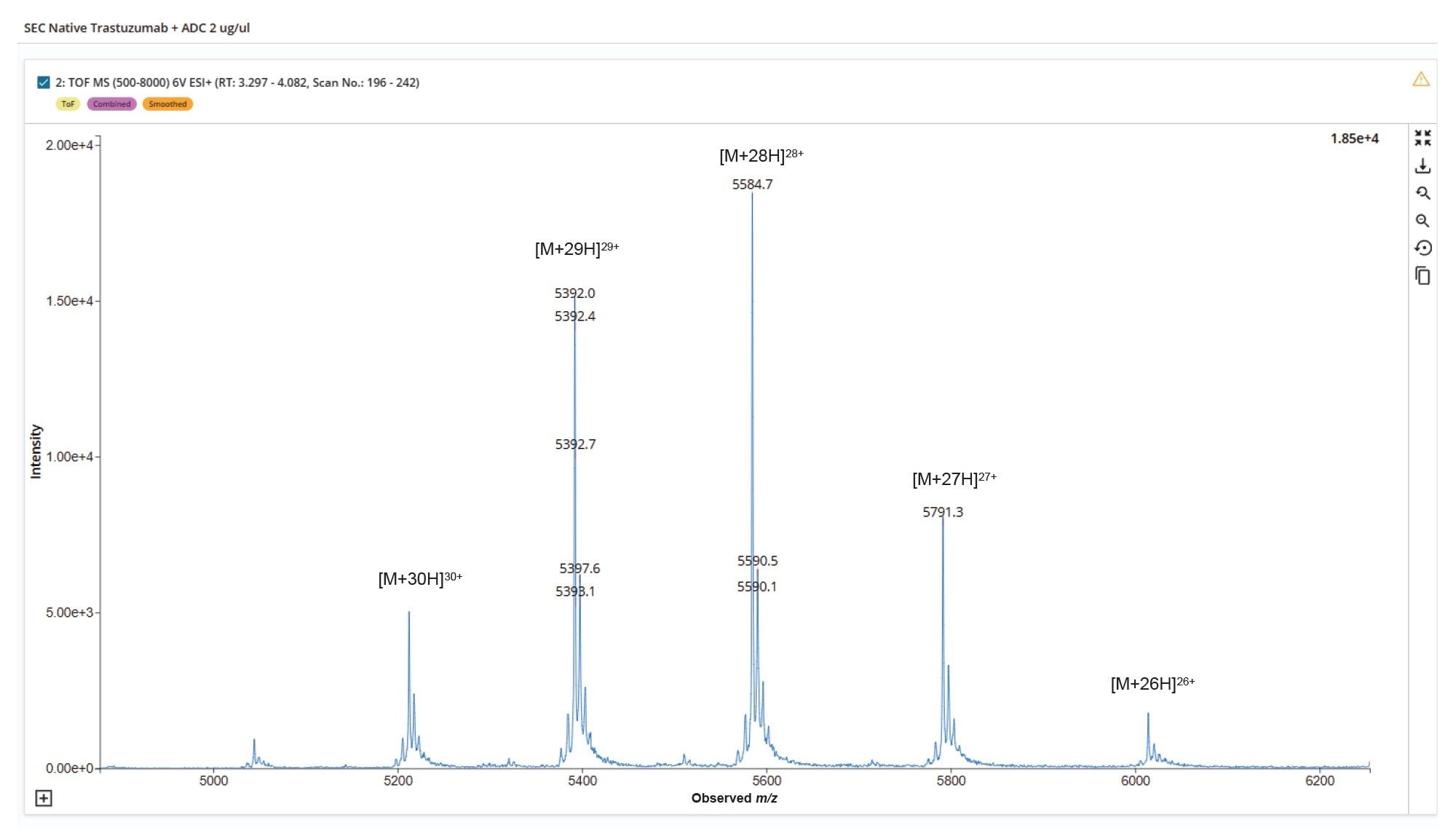
ENHERTU (fam-trastuzumab deruxtecan-nxki) is a commercial cysteine-conjugated ADC with a target DAR of approximately 8 molecules of cytotoxic agents per antibody.5 It is approved for the treatment of various cancers, including breast, gastric, and gastroesophageal malignancies.6 To enable intact ADC characterization by liquid chromatography mass spectrometry (LC-MS), non-denaturing separation conditions are required, particularly for cysteine-conjugated ADCs that may lack interchain disulfide bonds between heavy and light chains. Accordingly, a native SEC-MS approach was employed to analyze ENHERTU (fam-trastuzumab deruxtecan-nxki) in its intact form.
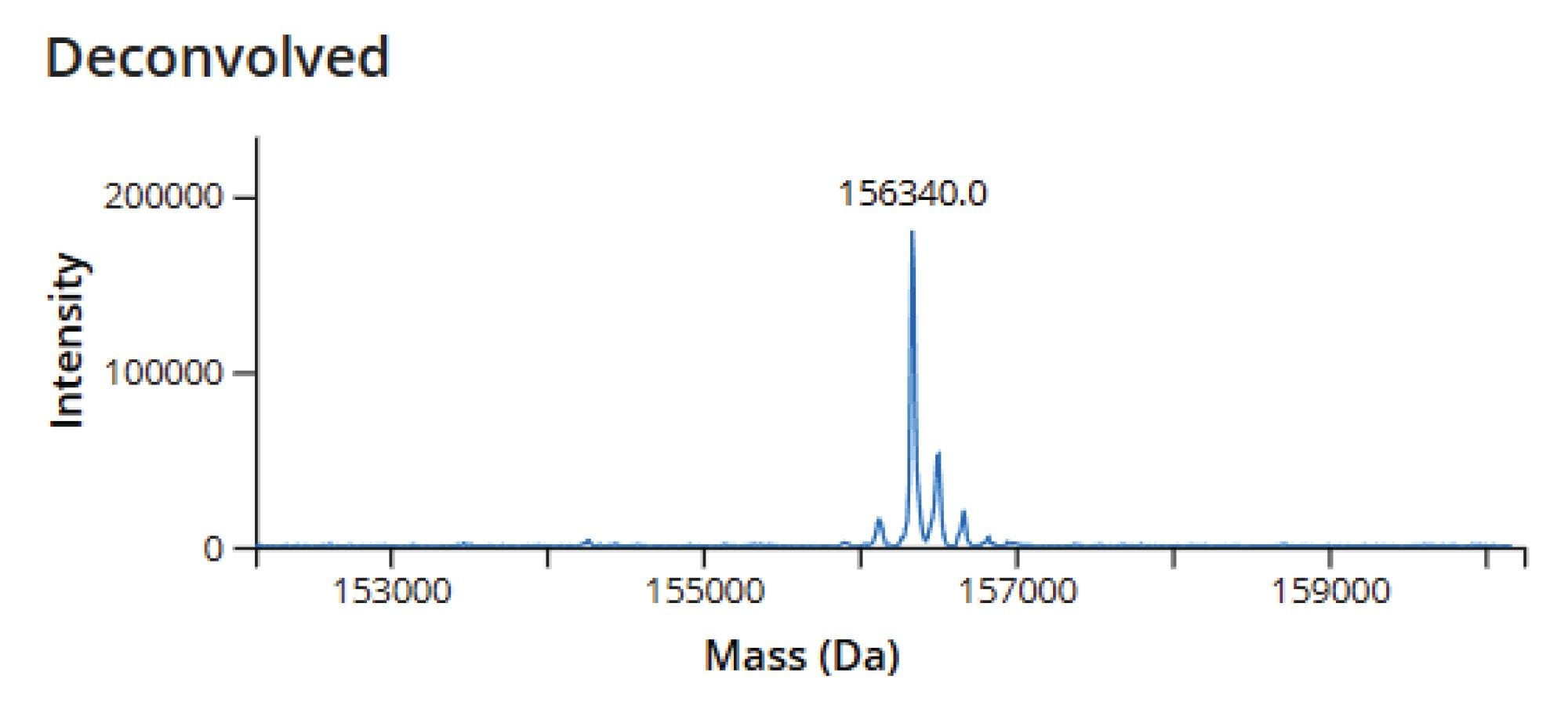
The native SEC-MS analysis of the ADC sample demonstrated clear and consistent N-glycoform separation for each of the observed charge states in the summed raw MS spectrum (Figure 6). This enabled confident deconvolution of the conjugated mAb (Figure 7), where the predominant species corresponded to N-glycoforms observed with Herceptin (trastuzumab) primarily conjugated with eight deruxtecan payloads, designated as “D8”. A minor proportion of species with alternative drug loadings such as six deruxtecan payloads “D6”, were also identified and assigned in the results tab of the INTACT Mass Application (Figure 8, Left Side). The calculation results tab (Figure 8, Right Side) presents the calculated DAR and overall drug distribution. As anticipated, the DAR for ENHERTU (fam-trastuzumab deruxtecan-nxki) was determined to be 7.97, consistent with analyses performed using the UNIFI Application Intact Mass workflow and previously published reports.5–8
Conclusion
The analysis of ADCs and other conjugates presents complexity and other challenges not encountered with the unmodified protein. In this study, the use of SEC-Native MS enabled analysis of the intact conjugate, despite loss of bridging disulfides in the antibody. The Xevo MRT Mass Spectrometer’s native MS mass range and exceptional mass accuracy capabilities delivered high quality data for both N-glycoform profiling measurements and ADC DAR and distribution measurements. The automation of these calculations by the waters_connect platform, and INTACT Mass App streamlines ADC analysis, provided consistent and confident results that can be utilized by analysts with a wide range of LC-MS experience across functions from characterization in early development to QC assays for product release.
References
- Shion H, Boyce P, Berger SJ, Yu YQ. INTACT Mass- a Versatile waters_connect Application for Rapid Mass Confirmation and Purity Assessment of Biotherapeutics. Waters Application Note 720007547. February 2022.
- Doneanu CE, Boyce P, Shion H, Fredette J, Berger SJ, Gastall H, Yu YQ. LC-MS Analysis of siRNA, Single Guide RNA and Impurities Using the BioAccord System with ACQUITY Premier and New Automated INTACT Mass Application. Waters Application Note 720007546. April 2022.
- D’Esposito RJ, Doneanu CE, Gastall H, Berger SJ, Yu YQ. RNA CQA Analysis using the BioAccord LC-MS System and INTACT Mass waters_connect Application. Waters Application Note 720008130. November 2023.
- Ranbaduge N, Shion H, Yu YQ. Streamlined LC-MS Analysis of Stress Induced Impurities of a Synthetic Peptide using the BioAccord System and the waters_connect INTACT Mass Application. Waters Application Note 720007752. October 2022.
- Daiichi Sankyo, Inc. (2025). Mechanism of Action | ENHERTU® (fam-trastuzumab deruxtecan-nxki). https://www.enhertuhcp.com/en/mechanism-of-action
- Riccardi F, Bo MD, Macor P, Toffoli G. A comprehensive overview on antibody-drug conjugates: from the conceptualization to cancer therapy. Front. Pharmacol. 14. 2023.
- Shion H, Yu YQ, Chen W. Analysis of Antibody Drug Conjugates (ADCs) by Native Mass Spectrometry on the BioAccord System. Waters Application Note 720006570. May 2019.
- Ippoliti, S., Yu, Y. Q., & Berger, S. (2025). The INTACT Mass Application in waters_connect™ Platform Streamlines ADC DAR and Drug Distribution Analysis. Waters Application Note 720008818. October 2025.
720009175, December 2025