The Achievability of Single Cell Lipidomics Utilizing the Novel Xevo™ MRT Mass Spectrometer
Abstract
We demonstrate the achievability of single cell analysis utilizing the novel Xevo™ MRT Mass Spectrometer coupled with an ACQUITY™ Premier LC System. Cultured THP-1 cells were lysed and extracted with IPA (100%) and diluted by five orders of magnitude. Using Lipostar (Mass Analytica, Barcelona, Spain) for peak picking and lipid identification, up to 180 lipids could be identified from the equivalence of a THP-1 single cell using stringent curation, providing highly confident identifications when utilising the Lipid Maps database (LMSD). The Xevo MRT MS demonstrates high sensitivity, and consistent sub-ppm mass accuracy, enabling highly confident lipid identifications, and thus providing a robust solution for single cell omics.
Benefits
- Dynamic range of five orders of magnitude enabling analysis of complex samples with analytes at varying concentrations
- High levels of sensitivity, allowing detection at single cell levels when utilizing standard flow chromatography
- Excellent data reproducibility facilitating statistical analyses, allowing for confident biological interpretation
- Consistent sub 1 ppm mass accuracy with <500 ppb RMS for confident identifications
- Resulting data can be simultaneously exported in mzML format enabling seamless integration with third party applications
Introduction
The development of cutting-edge mass spectrometry with high levels of sensitivity has led to single-cell OMICs gaining significant momentum in recent years. The importance of this is due to recent revelations that morphologically and even genetically identical cells exhibit profound heterogeneity on a transcriptomic, proteomic, metabolomic, and lipidomic level. Thus, to probe into population heterogeneity, single cell analysis is becoming increasingly vital to answer previously unanswerable biological questions. However, unlike transcriptomic approaches which rely on signal amplification, mass spectrometry based lipidomic or metabolomic approaches are still in their relative infancy.
This work assess the feasibility of single cell lipidomics on the Xevo MRT MS, that has shown to deliver excellent dynamic range, sensitivity, and mass accuracy, which are shown to deliver excellent dynamic range, sensitivity, and mass accuracy, for accurate identifications of lipids in single cell MS experiments.
Experimental
Sample Preparation
THP-1 was cultivated in a 75 mL flask containing RPMI-1640, 10% foetal bovine serum (FBS), 1% Non-Essential Amino Acids, 1% Sodium Pyruvate, 1% Glutamax, and 1% Kanamycin and was incubated at 37 °C and 5% CO2. THP-1 cells (20 µL) were collected and added to trypan blue (0.4%, 20 µL). These were counted with a disposable hemacytometer (C-CHIP DHC-N01, NanoEntek). The cells were harvested by centrifugation and washed with DPBS twice prior to being diluted to a density of 1x106/ mL. The cells were centrifuged, and the supernatant was removed, before centrifugation and removal of residual DPBS.
The bulk THP-1 cells were extracted in IPA containing 100 ng/ml EquiSPLASH™ (Avanti Lipids) to a final concentration of 5,000 cells/µL. This was shaken on a shaker set to 800 RPM, 5 °C, for one hour. The pellet was vortexed and returned to the shaker for a further hour before being centrifuged at 12,000 RPM for six minutes and the supernatant transferred to a TruView™ LCMS total recovery vial (Waters, p/n: 186005669CV). The sample was then further diluted to a concentration of 500, 50, 5, and 0.5 cells/µL.
Liquid Chromatography (LC)
LC analysis was performed using an ACQUITY Premier LC System equipped with a binary solvent manager, sample manager and column heater. Chromatographic separations were performed on an Waters ACQUITY Premier UPLC™ C18 CSH™, 1.7 µm, 2.1 x 100 mm (p/n: 186009461). The mobile phase was composed of 60:39:1 ACN:Water:1M ammonium formate +0.1% Formic Acid (A) and 90:9:1 IPA:ACN:1M ammonium formate +0.1% formic acid (B) with an active gradient of 10 minutes and total run time of 12 minutes. The sample manager was set to 8 °C, and the column heater was set to 55 °C. The column was directly attached to the probe rather than via the switching valve. 2 µL of the THP-1 cell dilution was injected onto the column.
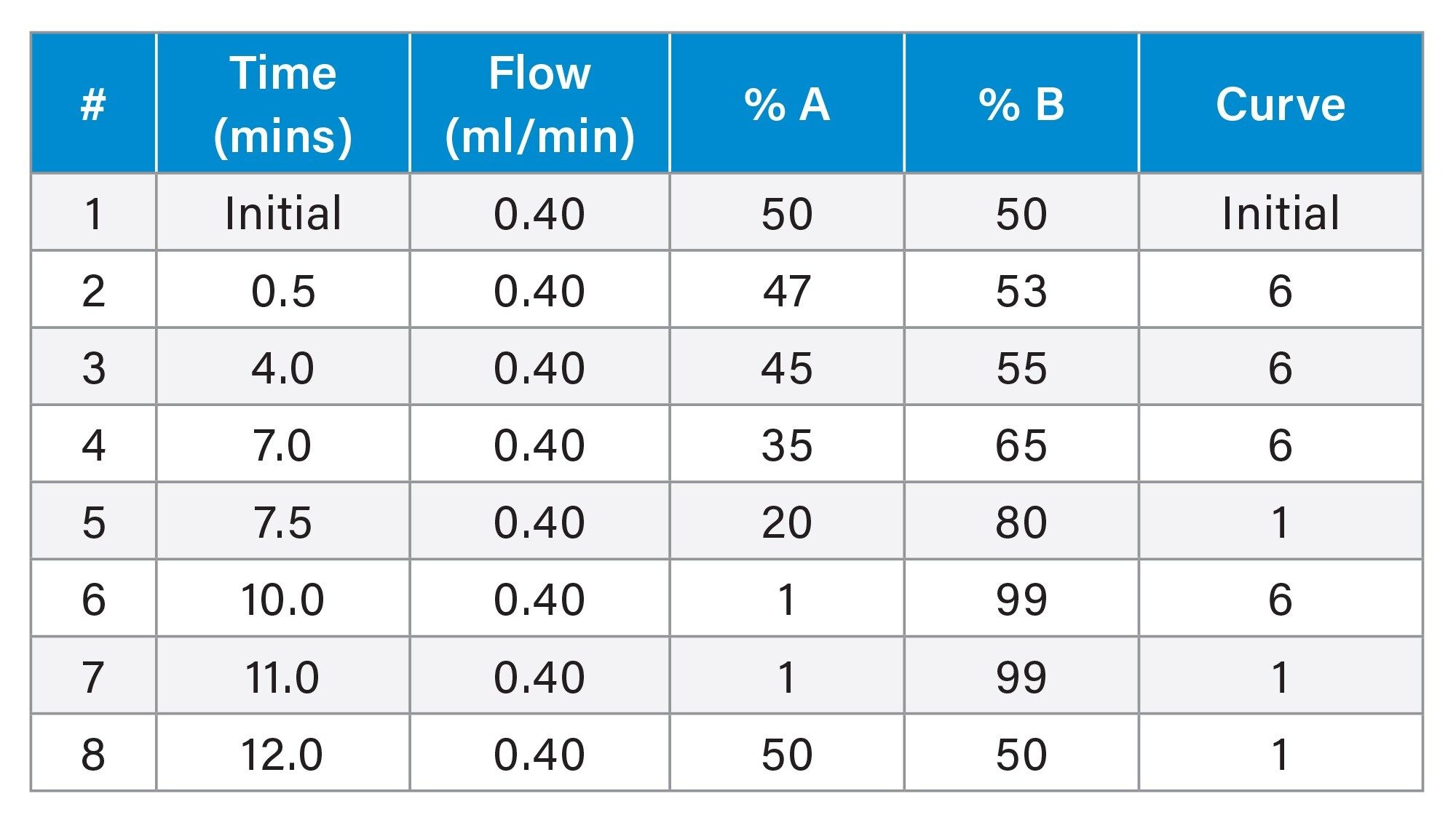
Mass Spectrometry (MS)
MS analysis was performed using the Xevo MRT MS. The MS was operated with electrospray ionisation in positive (ESI+) ion mode. The capillary voltage was set to 2.8 kV, cone voltage 40 V, source temperature 120 °C, desolvation temperature 500 °C, with a cone gas (N2) flow rate of 50 L/hr and desolvation gas flow rate of 750 L/hr. The acquisition rate was 10 Hz, and data acquired as MSE continuum, with the quadrupole operating in manual profile (400, 600, 850 m/z). Lockmass (leucine enkephalin, m/z 556.27658) was delivered via an external pump at 10 µL/min.

Data Analysis
The mzML files were imported into Lipostar (v 2.1.7b3 x64) with a peak picking threshold of 20,000 and 45,000 for MS and MS2 respectively. The data were searched against either the in-house Waters™ database, or the Lipid Maps database (LMSD) using default settings for scoring, and non-identified compounds were filtered out of the analysis. The resulting compound measurements were exported prior to being imported into MetaboAnalyst for additional statistical analysis and data visualisation. 5
Results and Discussion
Cells were extracted in triplicate with IPA containing 100 ng/ml EquiSPLASH and were diluted over 5 orders of magnitude (1, 10, 100, 1000, 10,000 cells per injection). Blanks (100% IPA) and EquiSPLASH (100 ng/ml), were also prepared. These were analysed on a Xevo MRT MS coupled to an ACQUITY Premier UPLC operating on the waters_connect™ Software Platform as described in the experimental. The data were exported in mzML format and processed using Lipostar with both an in-house database and the lipid maps database (LMSD).
The rapid 12-minute LC method is previously described in A Robust and Reproducible Reversed-Phase Lipid Profiling Method for Large Sample Sets | Waters (Plumb et al., 2020). Coupled with MSE acquisition at 10 Hz on the Xevo MRT MS this method provided impressive sensitivity at a single cell level. Furthermore, with mzML export written into the acquisition method editor, data could easily be transferred to and analysed on various third party applications. Data acquired on the Xevo MRT can be directly accessed by Lipostar through the API client configuration, or alternatively mzML files can be imported as described in this work.
Using Lipostar, data were processed with a blank filter, and variables were annotated using the database of choice. Lipostar enables users to filter lipid annotations according to the overall confidence score which is composed of the weighted average of accurate mass, isotope pattern and fragmentation.
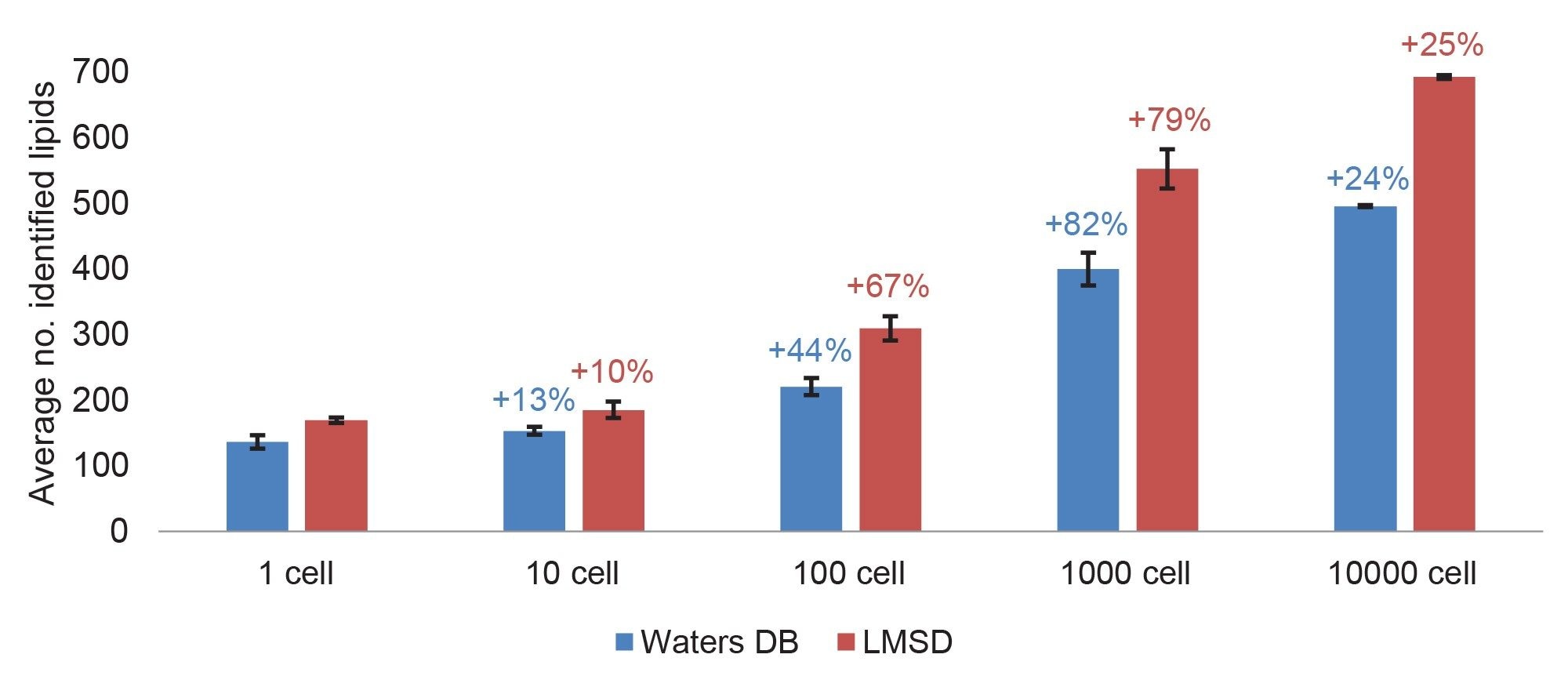
The resulting identifications with a high confidence score were exported, and lipids identified from the standard sample (EquiSPLASH 100 ng/ml) with a peak area > peak area in 1-cell were removed. The average number of identified curated lipids for the triplicate injections of THP-1 extracted cells ranged from 496 (in-house DB)/692 (LMSD) for 10,000 cells to 136 (in-house DB)/169 (LMSD) for a single cell (Figure 1).
Both databases resulted in the largest increase in identifications between 100–1000 cells (79–82% increase). This suggests that having a bulk sample of 1000 cells is adequate to create a database for this cell line and complements lipid identification for more diluted samples such as a single cell.
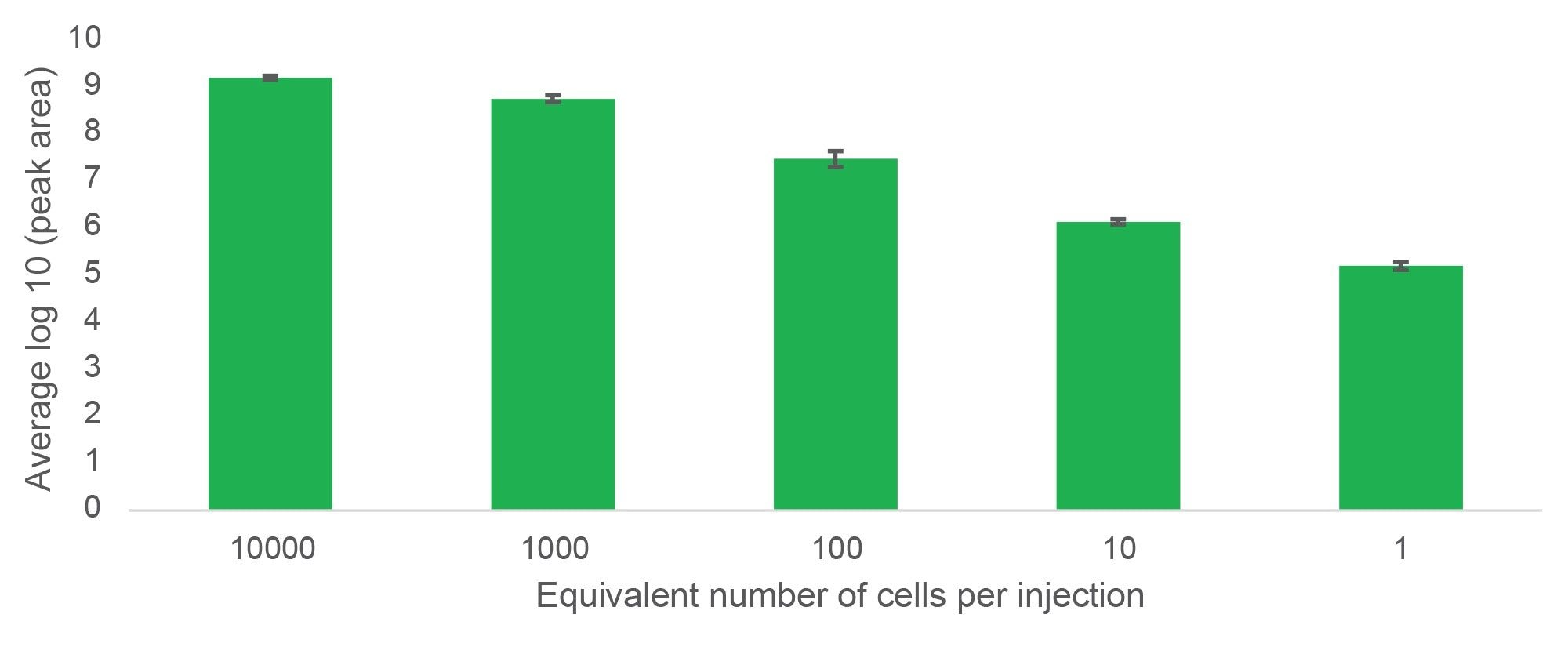
Lipids could be tracked over five orders of magnitude from 10,000 cells to a single cell equivalent, thus demonstrating the extended dynamic range of the Xevo MRT MS. This can be visualised with the lipid PC (36:2) in Figure 2.
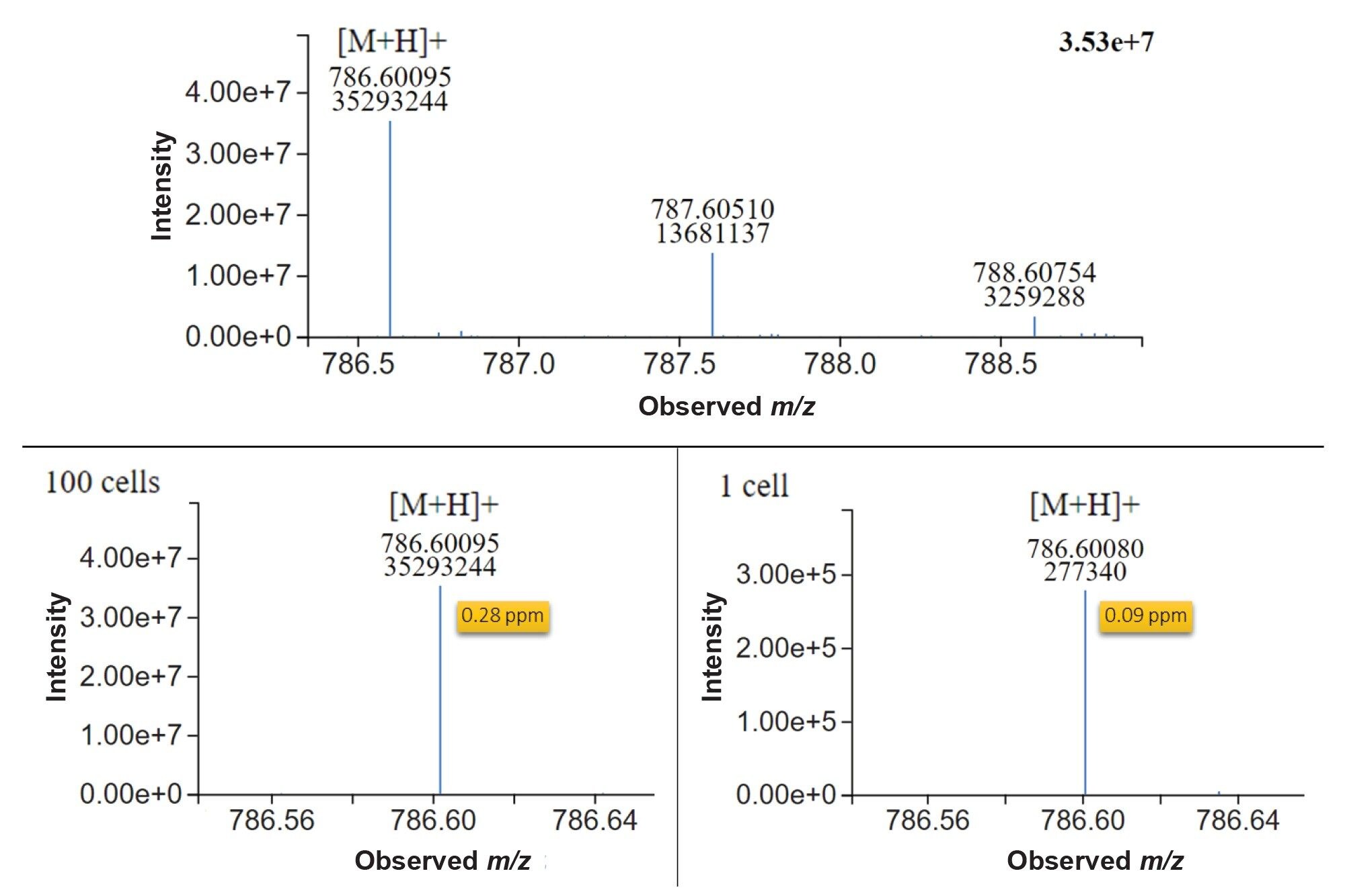
Raw MS1 spectra visualised in LC-MS toolkit application showed clearly defined peaks both at 100 cells and single cell levels (Figure 3). Furthermore, mass accuracy was consistently sub 500 ppb, providing robust and reliable identification of this lipid. The excellent mass accuracy achievable at such low concentrations with the Xevo MRT MS vastly reduces the number of tentative annotations, and combined with MS2 fragmentation data, enables analysts to be confident in the identifications returned.
Fragment ions and database matching of identified lipids could be further interrogated with Lipostar. For example, the feature 768.5521@4.04 was identified in the Lipostar software with a confidence level of three stars (74.33) as the [M+Na]+ adduct of the lipid PE(36:1). The MS2 spectrum was identified with two matching fragment ions (627.533, and 341.304), and additional probing of this spectrum using in silico fragmentation with MS/MS validator identified a further 11 fragments matching to this compound as displayed in Figure 4.
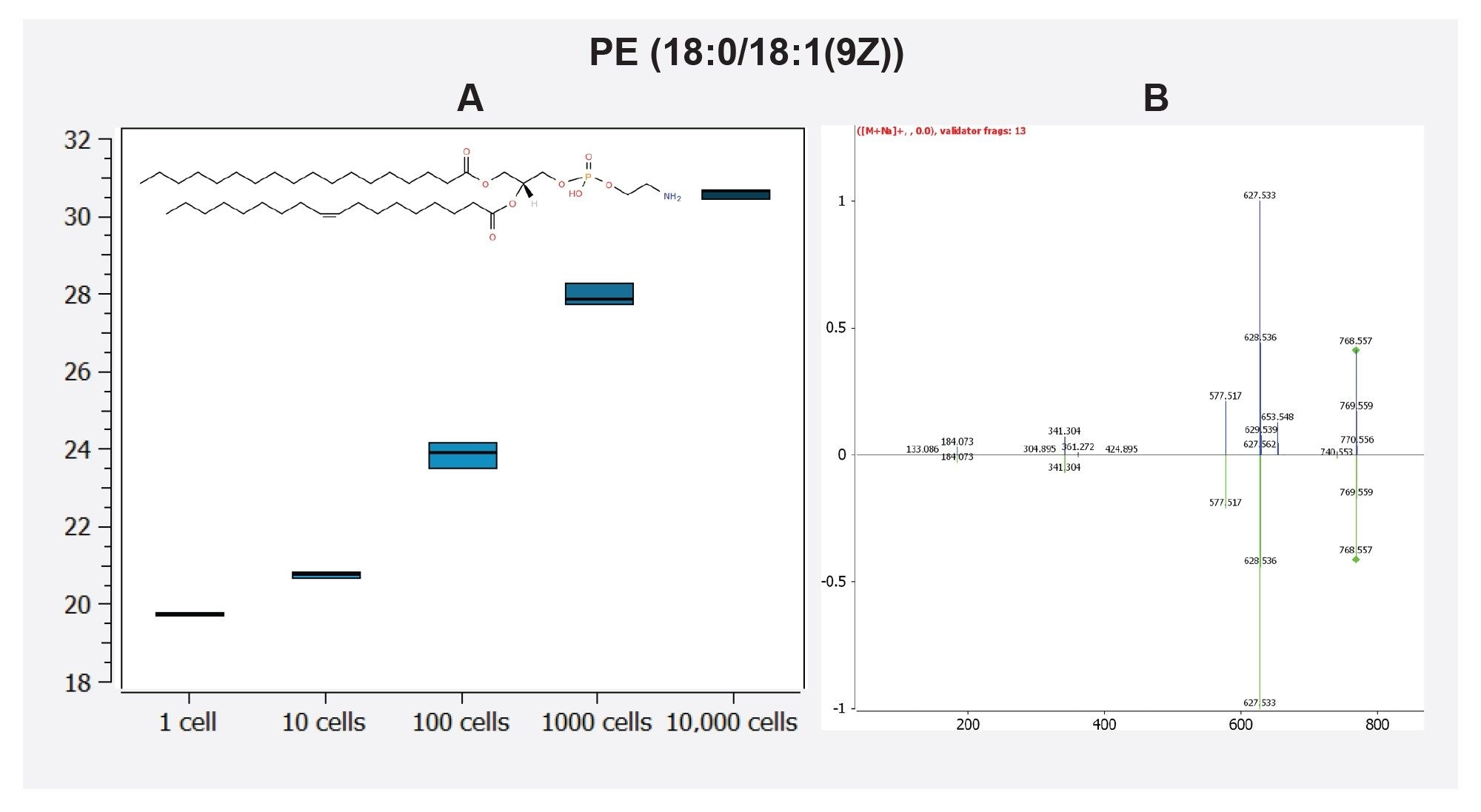
Conclusion
We demonstrate that the Xevo MRT Mass Spectrometer coupled with an ACQUITY Premier LC is capable of single cell mass spectrometry with greater than 150 lipids routinely identified from samples equivalent to a single cell. Furthermore, the excellent mass accuracy of less than 500 ppb provides confidence in lipid identifications particularly in combination with Lipostar (Mass Analytica) which provides a seamless and robust pipeline for annotation and identification of lipids. This is highlighted with PC (36:2) and PE (36:1) in Figure 3 and 4 respectively. Ultimately, the Xevo MRT MS demonstrates sensitivity over five orders of magnitude, and paired with Lipostar, represents an excellent solution for single cell lipidomic analyses.
References
- C. Seydel, Nat Methods, 2021, 18, 1452–1456.
- B. Carter and K. Zhao, Nat Rev Genet, 2021, 22, 235–250.
- C. E. Randolph, P. Manchanda, H. Arora, S. Iyer, P. Saklani, C. Beveridge, and G. Chopra, TrAC Trends in Analytical Chemistry, 2023, 169, 117350.
- G. Liebisch, E. Fahy, J. Aoki, E. A. Dennis, T. Durand, C. S. Ejsing, M. Fedorova, I. Feussner, W. J. Griffiths, H. Köfeler, A. H. Merrill, R. C. Murphy, V. B. O’Donnell, O. Oskolkova, S. Subramaniam, M. J. O. Wakelam, and F. Spener, J Lipid Res, 2020, 61, 1539–1555.
- Z. Pang, L. Xu, C. Viau, Y. Lu, R. Salavati, N. Basu, and J. Xia, Nat Commun, 2024, 15, 3675.
- L. Goracci, P. Tiberi, S. Di Bona, S. Bonciarelli, G. I. Passeri, M. Piroddi, S. Moretti, C. Volpi, I. Zamora, and G. Cruciani, Anal Chem, 2024, 96, 1468–1477.
720008569, October 2024