An Automated Sample Preparation Protocol to Determine the Encapsulation Efficiency of RNA-Loaded Lipid Nanoparticles Using Andrew+ Pipetting Robot

Abstract
The determination of encapsulation efficiencies in RNA loaded lipid nanoparticles is often performed manually, which is a time-consuming and laborious process in a routine or routine-like environment. The following work outlines the development of an automated procedure using the Andrew+ Pipetting Robot.
Benefits
- Automated execution of a RiboGreen assay to determine mRNA-LNP encapsulation efficiencies
- A high-throughput protocol with enhanced flexibility is provided to enable the analysis of up to 16 samples at a reduced assay time (from 120 min to 45 min)
- Encapsulation efficiencies are comparable to manual pipetting results
- Routine analytical platform is supposed to support high-throughput RNA formulation development settings
Introduction
mRNA delivery via non-viral vectors, particularly lipid nanoparticles (LNPs), has gained momentum following the successful use of LNPs in COVID-19 vaccines.1,2 LNPs typically comprise four lipid components—an ionizable lipid, DSPC (1,2-distearoyl-sn-glycero-3-phosphocholine), cholesterol, and PEGylated lipid each contributing specific functionality.3,4 These nanoparticles effectively deliver RNA molecules, which can be rapidly designed to address various medical challenges that include infectious diseases, cancer, and heart diseases, positioning LNPs as a promising vehicle for the delivery of RNA therapeutics.
The LNP optimization process typically involves fine-tuning the lipid components to enhance stability, efficacy, and safety and even enable selective organ targeting for more precise delivery.6 To ensure the quality of mRNA vaccines, guidelines from the USP recommend assessing critical quality attributes, including the degree of encapsulation, to guarantee accurate dosing.7
The RiboGreen assay is a highly sensitive fluorescence-based method for quantifying RNA and determining encapsulation efficiency (EE), offering specificity against other matrix components, in contrast to UV detection.8 While this assay is routinely used in formulation development laboratories, its manual execution can be cumbersome and time-consuming in high-throughput settings. Therefore, the objective of this work was to establish an automated protocol to enhance the in-house high-throughput formulation development platform. The resulting fully automated protocol enables the analysis of up to 16 samples in under 60 minutes, enabling reproducibility across laboratories spanning from R&D to CMC sites within pharmaceutical companies.
Experimental
Encapsulation Efficiency
To calculate the LNP encapsulation efficiency and RNA concentration, a Quant-iT RiboGreen® RNA assay (Thermo Scientific) was performed according to the manufacturer’s instructions. The provided TE-Buffer (10 mM Tris-HCl pH 8, 0.1 mM EDTA) was diluted (1:20, v/v) in nuclease free water prior use. The RiboGreen solution was prepared freshly by diluting the RiboGreen reagent (1:166, v/v) in TE Buffer. Triton™ X-100 was purchased from Thermo Scientific , and was diluted (1:50, v/v) to prepare the TX-Buffer.
Lipid nanoparticles (LNPs) containing mRNA were prepared in-house following established protocols.8,9 The formulated samples were diluted to a concentration of 50–75 µg/mL, thus prepared to result in a final concentration of 417–625 ng/mL for quantitation.
An mRNA (proprietary) stock solution (1 mg/mL) was used to prepare the calibrant stock solution. Next, 10 µL of the stock solution was diluted 1:10 (v/v) in TE buffer and the RNA concentration was measured by the Nanodrop (2 x 2 µL), which was used to further dilute the sample to a final concentration of 5 µg/mL (±0.5 µg/mL), to ensure the accurate quantification.
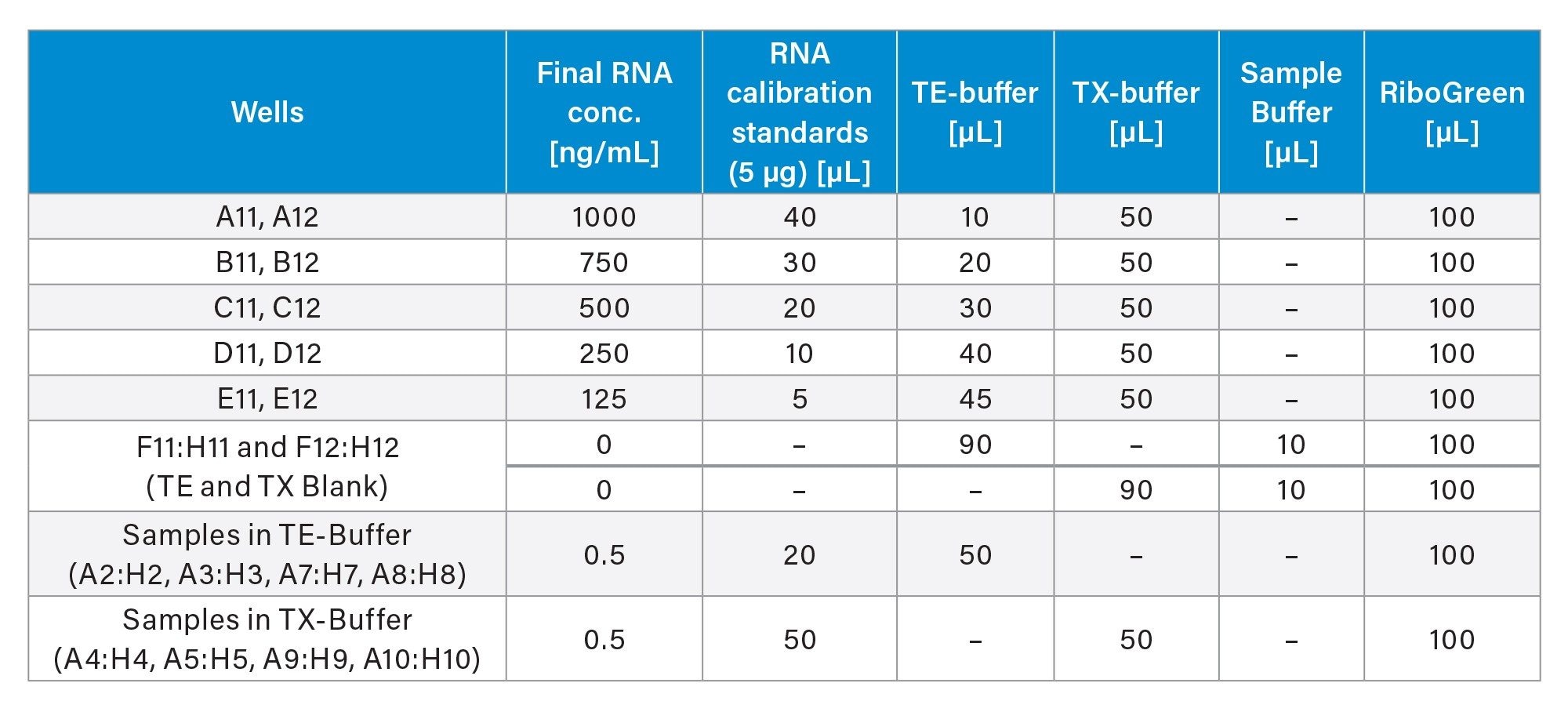
The encapsulation efficiency was calculated by subtracting the non-encapsulated RNA concentration (in TE buffer) from the total RNA concentration (in TX buffer) after LNP deformulation. A five-point calibration curve (average of duplicates) was employed in TX buffer to account for the intrinsic fluorescence of the TX buffer. Sample measurements were corrected by subtracting average blank values. The pipetting scheme is summarized in Table 1.
Automated Liquid Handling
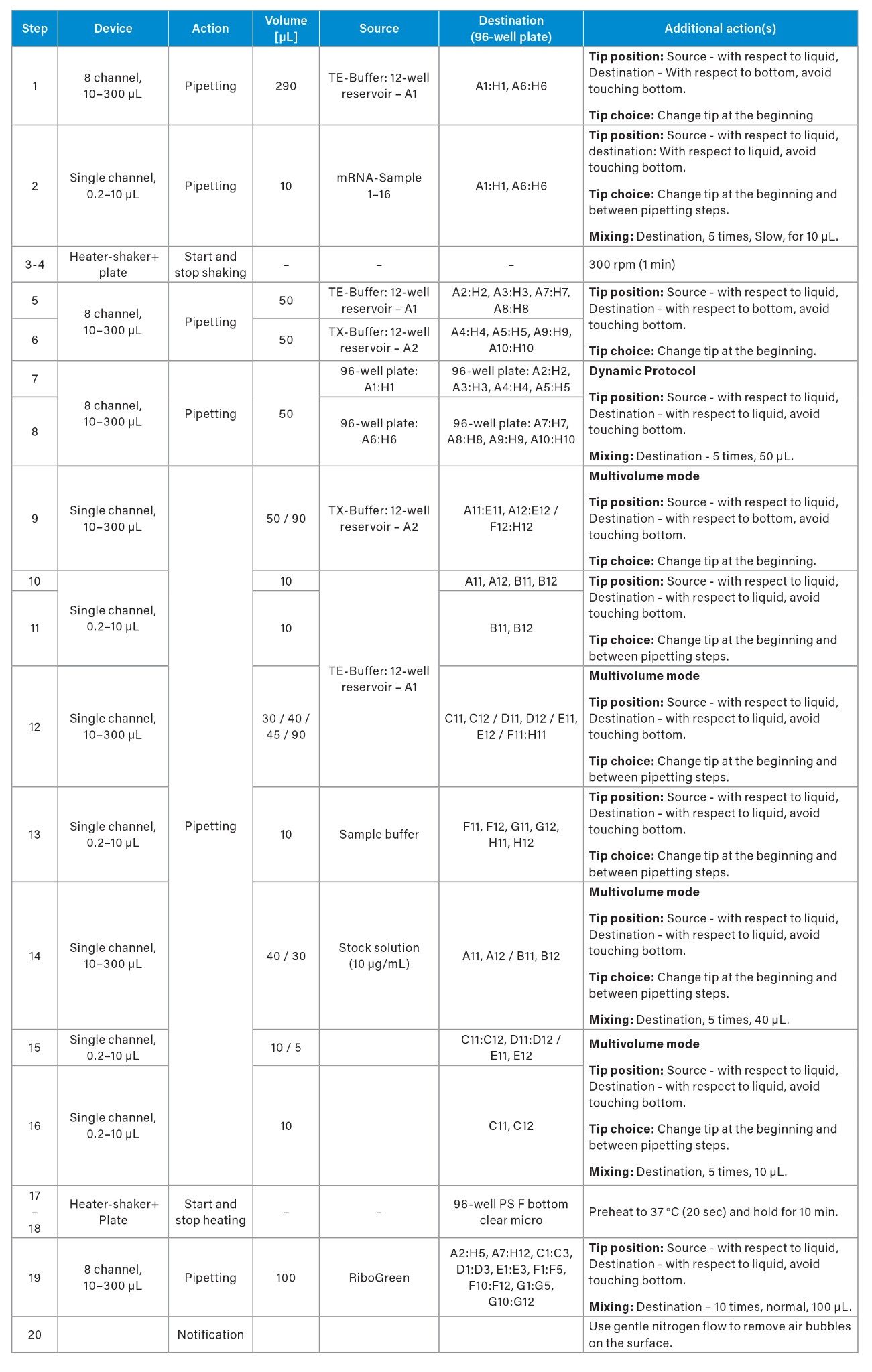
Automated liquid handling steps were performed using the Andrew+ Pipetting Robot (Figure 1) , using a protocol developed and executed using OneLab™ Software equipped with a 10 µL and 300 µL single channel pipette and a Heater-Shaker™ Plate (Waters Corporation). The setup further comprised the corresponding dominos for the following labware: Two Tip Insertion Systems, 50 mL Conical Centrifuge Tube Domino, Microtube Domino (Waters Corporation) with 2 mL Eppendorf Safe-Lock tubes (Eppenforf) , Axygen® 12-well reservoir (16 mL), and a 96-well PS F-bottom clear microplate (Greiner, Frickenhausen) used as the target labware.

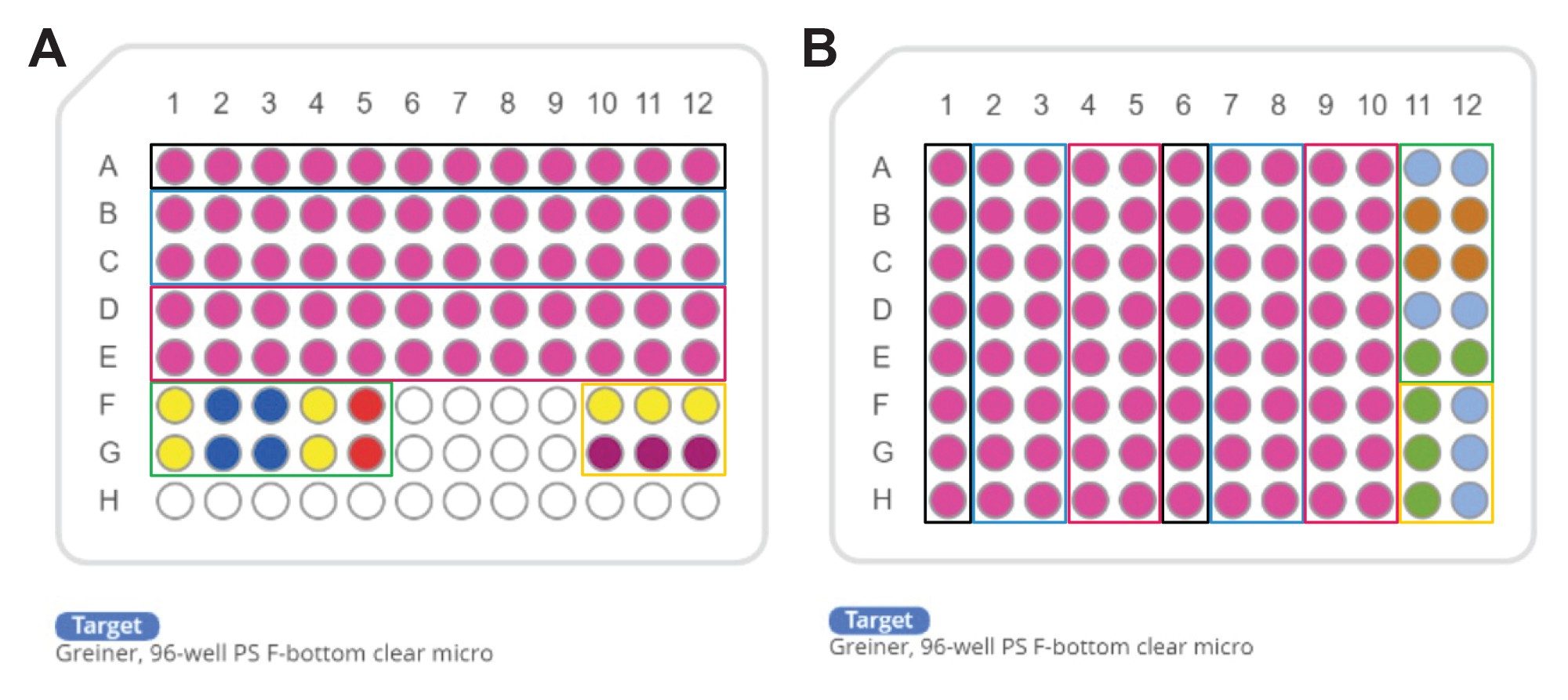
Two pipetting schemes, 12 samples (Figure 2A) and 16 samples (Figure 2B), were implemented. The 12-sample protocol, adapted from Invitrogen, is currently in use within our laboratory. The 16-sample scheme was developed from an optimization process, which is detailed in the following sections.
Results and Discussion
Manual Liquid Handling
The RiboGreen assay is often performed manually, since lab automation is an ever-progressing field. The flexibility of manual pipetting offers several advantages, such as the use of a multichannel pipettes (e.g. 8-channel) as an n-channel (n≤8) pipette for a reduced number of samples, for example 4 samples could be processed by only using 4 of 8 channels (n=4) in parallel. In general, this allows for efficient and time-saving pipetting in a manual workflow. However, manual pipetting also comes with its disadvantages. One challenge is the consistent attachment of pipette tips to the multi-pipette, as some tips may not adhere properly. Additionally, manual pipetting requires a high level of care to ensure accurate and precise results, making it a time-consuming process and increasing the risk of human error.10,11 Thus, manual pipetting offers certain advantages in terms of flexibility and cost-effectiveness but also presents challenges that can impact the accuracy and efficiency of pipetting procedures.
Automated Liquid Handling
Automated pipetting systems present a myriad of advantages that significantly enhance both the efficiency and accuracy of the RiboGreen assay. Particularly when working with surfactants such as Triton, a gentle reproducible mixing speed is advantageous, since the formation of bubbles is a known disadvantage to pipetting accuracy. Therefore, during the addition of samples to the wells containing TX-buffer (step 7 and 8) a gentle pipetting mixing step was employed. The utilization of reverse pipetting could further decrease the likelihood of bubble formation but was not implemented in this protocol. During pipetting to the wells containing TE-buffer (step 7 and 8), the gentle incubation/mixing process is further critical to preserve the integrity and retain the quality of the sensitive LNPs.
To establish efficient protocols, several key considerations were implemented. The initial protocol (Figure 2A) was adapted directly from the Invitrogen method to an automated format, demonstrating comparable results between manual and automated pipetting (data not shown). To enhance protocol efficiency, the pipetting scheme was transposed from a horizontal (Figure 2A) to a vertical orientation (Figure 2B), enabling to process up to 16 samples. This modification allowed for full utilization of all 96 wells in the plate, increasing well-plate usage from 83% to 100%. Additionally, the optimized pipetting actions remained consistent across both protocols. The integration of an 8-channel pipetting capability significantly improved time efficiency, resulting in a reduction of pipetting steps from 31 to 20 and decreased the overall experiment duration by from 120 min to 45 min (63%) as estimated by OneLab Software.
The following features were implemented across both setups. The dynamic protocol functionality enabled the flexible processing of 1 to 16 samples, enhancing workflow adaptability in research environments where sample numbers can vary significantly. The multivolume mode feature further contributes to the time efficiency of the protocol, reducing the total number of steps by approximately 21% (from 38 to 30 in the vertical setup, Figure 2A). Additionally, the calculation of required reagent volumes is performed by the OneLab Software, which optimizes reagent usage, thereby conserving resources and reducing overall costs associated with the assay, which can be particularly significant in high-throughput settings.
Further, the tip loading process of the multichannel pipette is ensured by the consistent force distribution, reducing tip attachment issues commonly associated with manual techniques. Overall, the advantages of automated pipetting—including precise mixing, reduced error rates, and optimized reagent usage—enhance the reliability and reproducibility of the assay, fostering more robust outcomes.
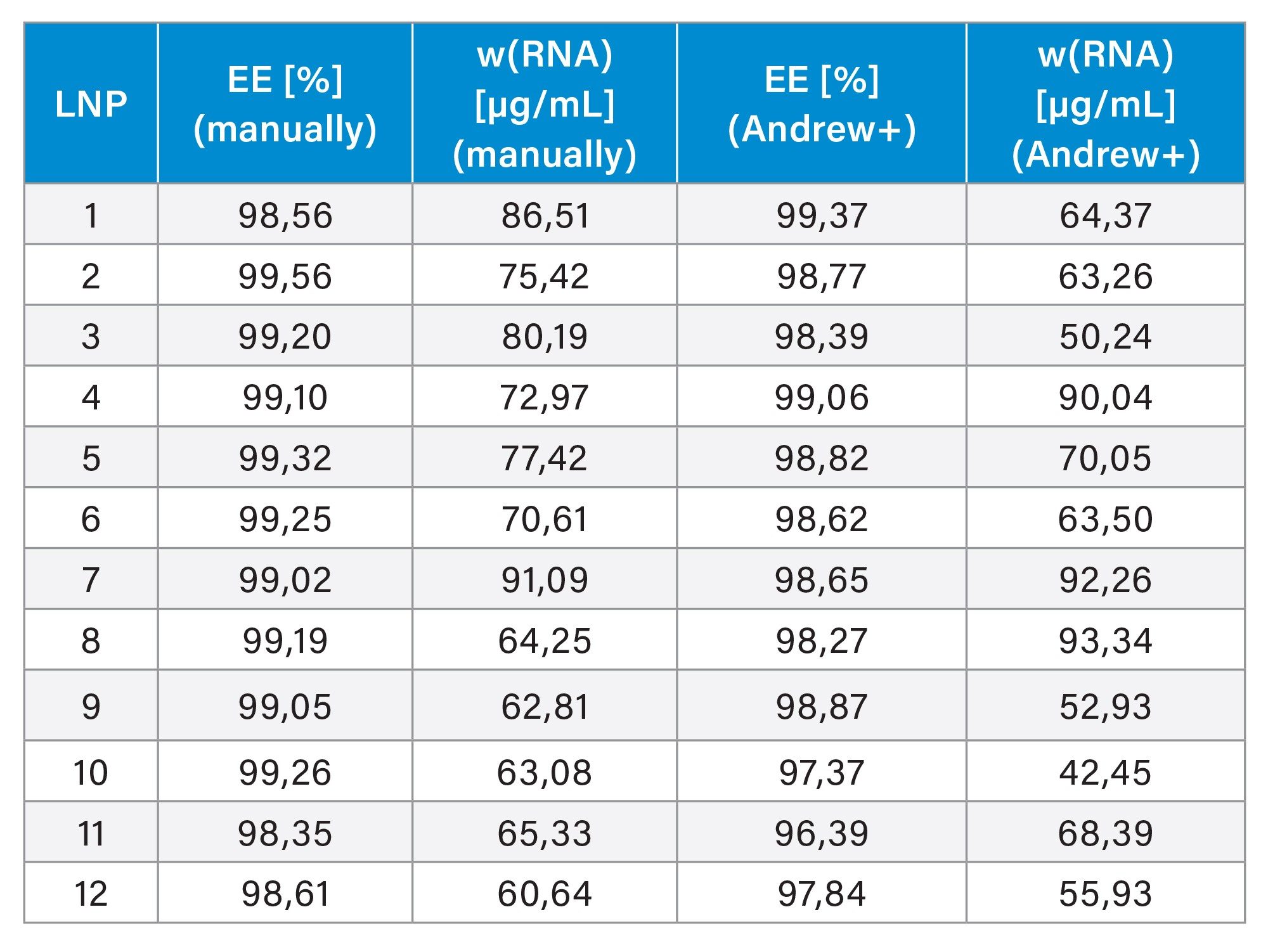
Encapsulation Efficiency and Concentration
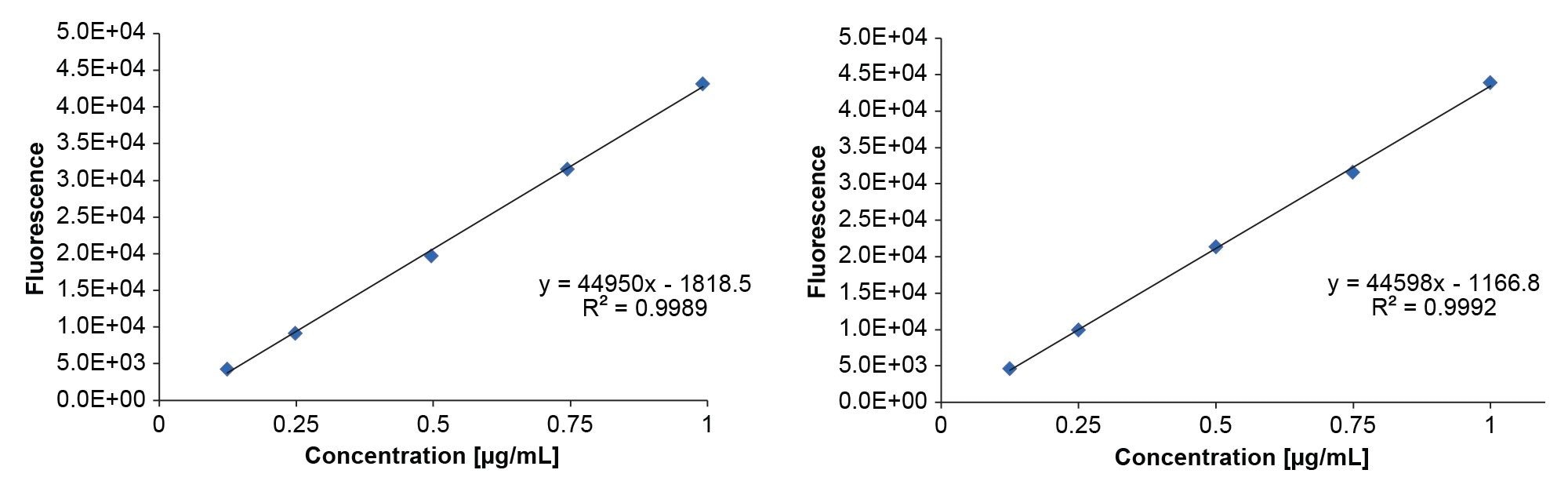
Twelve lipid nanoparticles (LNPs) were prepared for a study with varying amounts of DSPC (3–36%), which promotes the fusion with cell and endosomal membranes.5 The mRNA concentrations ranged between 50–75 µg/mL. The encapsulation efficiency was assessed using the RiboGreen assay, evaluating both the original non-automated protocol (12-samples) and the automated (16‑sample) protocol. A calibration curve was generated according to Table 1, resulting in sufficient linearity (R²>0.99) for both methods. All samples demonstrated high average encapsulation efficiency of 99.04 (RSD=0.36). Further, the automated assay was performed with a time offset of 30 days (storage at -80 °C) to assess the mRNA stability. The encapsulation efficiency was largely retained at an average of 98.37 (RSD=0.83). However, a decrease in mRNA concentration was observed after the storage period, that adds a freeze-thaw cycle to the samples, which is known to impair the stability of sensitive biological samples.12–15 Thus, differences in mRNA concentration measurements after storage are expected and have been observed in the past. It should be noted that the calibration curve is prepared with pure and intact mRNA to calibrate LNP samples, without matching the matrix, leading to potential inaccuracies.16,17 In conclusion, the automated assay, demonstrated performance that is at least comparable to manual pipetting.
The following considerations should be made to further simplify the workload for the user and increase flexibility. First, the calibration range can easily be adjusted by varying the RNA stock concentration solution to flexibly accommodate larger LNP concentrations, thus avoiding pre-dilution tasks before using the samples in the automated workflow. Secondly, to avoid pre-dilutions, varying initial LNP concentrations can be accounted for by using the multivolume mode to adjust the TE-buffer/LNP ratio in the dilution steps (step 1 and 2), across a defined LNP concentration range, e.g. between 10–100 µg/mL, that aligns with the calibration curve and the employed RiboGreen concentration. A 6-well reservoir could be used to accommodate larger required volumes of TX and TE buffers.
Conclusion
In conclusion, the commercial RiboGreen assay was fully automated on the Andrew+ Pipetting Robot. The 96-well plate setup maximized well-plate usage and utilized the 8-channel pipette, increasing throughput from 12 to 16 samples while reducing processing time from 120 minutes to 45 minutes. Protocol optimization involved the use of dynamic protocols and multivolume mode, enhancing flexibility and reducing the overall number of steps. Reliable mixing was implemented to address bubble production from Triton surfactant, which can impair fluorescence detection. Additionally, automatic calculation of required reagents, such as RiboGreen, helped reduce wastage of the dye. Comparisons between manual and automated pipetting demonstrated comparable encapsulation efficiencies, indicating that the provided protocol is well-suited for high-throughput laboratories conducting routine RiboGreen assays.
References
- R. Tenchov, R. Bird, A.E. Curtze, Q. Zhou, Lipid Nanoparticles─From Liposomes to mRNA Vaccine Delivery, a Landscape of Research Diversity and Advancement, ACS Nano 15(11) (2021) 16982–17015. https://doi.org/10.1021/acsnano.1c04996.
- K.S. Park, X. Sun, M.E. Aikins, J.J. Moon, Non-viral COVID-19 Vaccine Delivery Systems, Advanced Drug Delivery Reviews 169 (2021) 137–151. https://doi.org/10.1016/j.addr.2020.12.008.
- K.A. Hajj, K.A. Whitehead, Tools for Translation: Non-Viral Materials for Therapeutic mRNA Delivery, Nature Reviews Materials 2(10) (2017) 17056. https://doi.org/10.1038/natrevmats.2017.56.
- W. Li, F.C. Szoka, Lipid-based Nanoparticles for Nucleic Acid Delivery, Pharmaceutical Research 24(3) (2007) 438-449. https://doi.org/10.1007/s11095-006-9180-5.
- X. Cheng, R.J. Lee, The Role of Helper Lipids in Lipid Nanoparticles (LNPs) Designed for Oligonucleotide Delivery, Advanced Drug Delivery Reviews 99 (2016) 129–137. https://doi.org/10.1016/j.addr.2016.01.022.
- X. Wang, S. Liu, Y. Sun, X. Yu, S.M. Lee, Q. Cheng, T. Wei, J. Gong, J. Robinson, D. Zhang, X. Lian, P. Basak, D.J. Siegwart, Preparation of Selective Organ-Targeting (SORT) Lipid Nanoparticles (LNPs) Using Multiple Technical Methods for Tissue-Specific mRNA Delivery, Nature Protocols 18(1) (2023) 265–291. https://doi.org/10.1038/s41596-022-00755-x.
- USP, Analytical Procedures for Quality of mRNA Vaccines and Therapeutics, USP Vol. 3 (2024).
- L.J. Jones, S.T. Yue, C.-Y. Cheung, V.L. Singer, RNA Quantitation by Fluorescence-Based Solution Assay: RiboGreen Reagent Characterization, Analytical Biochemistry 265(2) (1998) 368–374. https://doi.org/10.1006/abio.1998.2914.
- N. Karekar, A. Reid Cahn, J. Morla-Folch, A. Saffon, R.W. Ward, A. Ananthanarayanan, A.J.P. Teunissen, N. Bhardwaj, N. Vabret, Protocol for the Development of mRNA Lipid Nanoparticle Vaccines and Analysis of Immunization Efficiency in Mice, STAR Protocols 5(2) (2024) 103087. https://doi.org/10.1016/j.xpro.2024.103087.
- I. Holland, J.A. Davies, Automation in the Life Science Research Laboratory, Frontiers in Bioengineering and Biotechnology 8 (2020). https://doi.org/10.3389/fbioe.2020.571777.
- M.M. Jessop-Fabre, N. Sonnenschein, Improving Reproducibility in Synthetic Biology, Frontiers in Bioengineering and Biotechnology 7 (2019). https://doi.org/10.3389/fbioe.2019.00018.
- K. Hashiba, M. Taguchi, S. Sakamoto, A. Otsu, Y. Maeda, H. Ebe, A. Okazaki, H. Harashima, Y. Sato, Overcoming Thermostability Challenges in mRNA–Lipid Nanoparticle Systems With Piperidine-based Ionizable Lipids, Communications Biology 7(1) (2024) 556. https://doi.org/10.1038/s42003-024-06235-0.
- R.E. Birdsall, D. Han, K. DeLaney, A. Kowalczyk, R. Cojocaru, M. Lauber, J.L. Huray, Monitoring Stability Indicating Impurities and Aldehyde Content in Lipid Nanoparticle Raw Material and Formulated Drugs, Journal of Chromatography B 1234 (2024) 124005. https://doi.org/10.1016/j.jchromb.2024.124005.
- M. Kamiya, M. Matsumoto, K. Yamashita, T. Izumi, M. Kawaguchi, S. Mizukami, M. Tsurumaru, H. Mukai, S. Kawakami, Stability Study of mRNA-Lipid Nanoparticles Exposed to Various Conditions Based on the Evaluation Between Physicochemical Properties and Their Relation with Protein Expression Ability, Pharmaceutics 14(11) (2022) 2357.
- R. Matthessen, R. Van Pottelberge, B. Goffin, G. De Winter, Impact of Mixing and Shaking on mRNA-LNP Drug Product Quality Characteristics, Scientific Reports 14(1) (2024) 19590. https://doi.org/10.1038/s41598-024-70680-4.
- M.S. Lowenthal, A.S. Antonishek, K.W. Phinney, Quantification of mRNA in Lipid Nanoparticles Using Mass Spectrometry, Analytical Chemistry (2024). https://doi.org/10.1021/acs.analchem.3c04406.
- G. Visconti, E. Olesti, V. González-Ruiz, G. Glauser, D. Tonoli, P. Lescuyer, N. Vuilleumier, S. Rudaz, Internal calibration as an Emerging Approach for Endogenous Analyte Quantification: Application to Steroids, Talanta 240 (2022) 123149. https://doi.org/10.1016/j.talanta.2021.123149.
Featured Products
720008732, March 2025