
This is an Application Brief and does not contain a detailed Experimental section.
This application brief demonstrates a streamlined workflow for a non-targeted analysis comparing the ingredients of fresh and dry Ginkgo leaves using an ACQUITY UPLC-SYNAPT G2 HDMS/MarkerLynx XS-based System.
The combination of UPLC separations and high resolution mass spectrometry with multivariate statistical analysis facilitates rapid and comprehensive profiling of complex samples such as Traditional Chinese Medicines.
UPLC/TOF MS-based multivariate statistical analysis is a proven solution for studying grouping patterns of complex samples. It has been widely accepted for the application of metabolomics to study the changes of endogenous ingredients from biological matrices before and after certain stimuli, such as administration of a drug, or before and after disease onset. This analytical strategy, however, has applicability for a much wider array of applications that require the comparison of multiple groups of complex samples. Specifically, in the field of Traditional Chinese Medicine (TCM), a common but challenging task is to compare different herbal extracts; either the same herb harvested at different times or locations, or stored under different conditions.
In this application example, a fresh ginkgo leaf extract is compared with a dry ginkgo leaf extract to determine if there is a natural grouping amongst the two and if so, to further identify the leading markers that differentiate the two sample groups. Typically, in TCM preparations, it is the dry leaves that are stored and used for treatment.
Ginkgo leaves were harvested from Liuyan, Hunan Province in China in 2009. A standardized extraction procedure was applied to the leaves to produce final extracts for LC-MS analysis. Samples were analyzed using and ACQUITY UPLC-SYANPT G2 HDMS System. An ACQUITY UPLC HSS T3 2.1 x 100 mm column was used, mobile phases were 5 mM ammonium acetate in water at pH 5.0 (mobile phase A) and methanol (mobile phase B). The sample run time was 22 minutes; each sample was injected no less than 3 times in negative ion electrospray mode. Data acquisition was performed using MSE, a novel method of data acquisition that allows both precursor and fragment ion data to be obtained on virtually every detectable component in the sample. Data processing was performed with the MarkerLynx XS Application Manager.
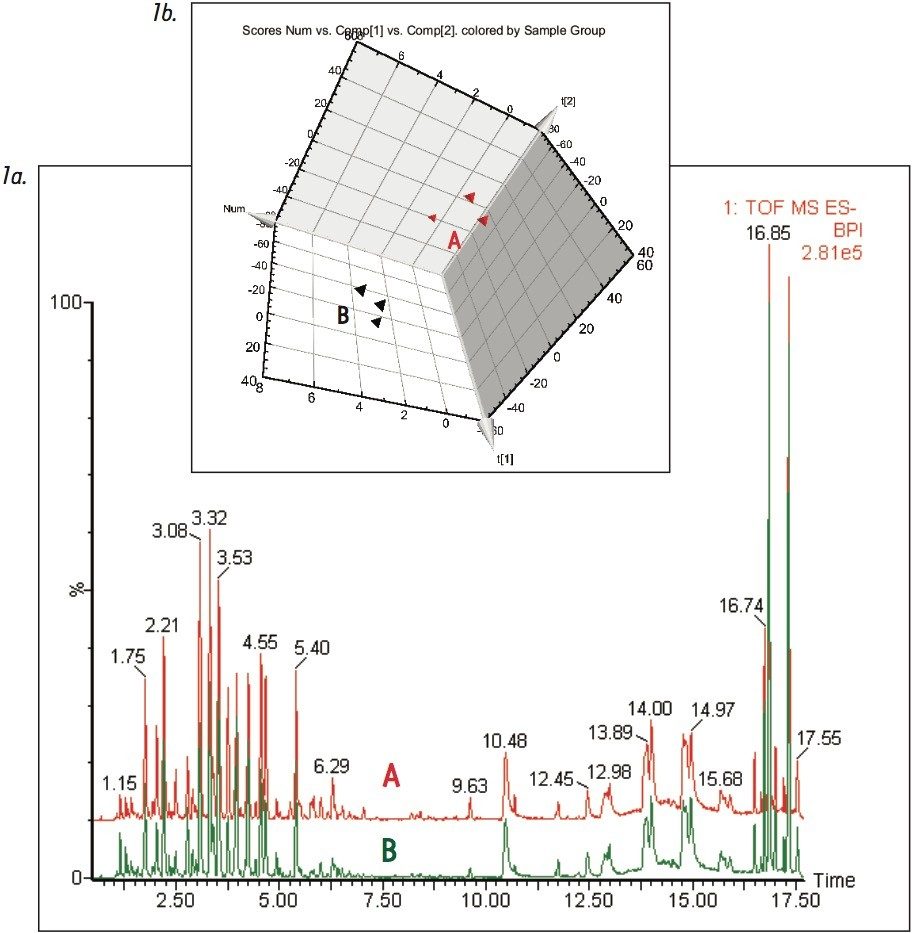
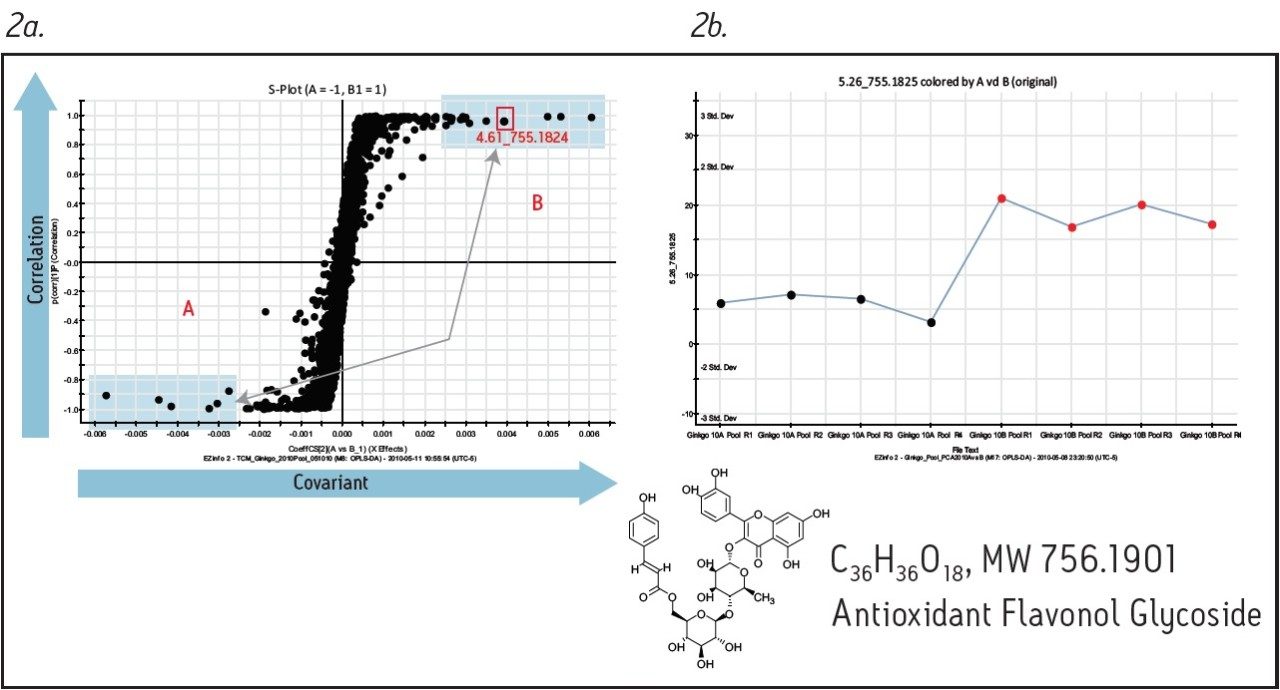
Figure 1 shows the base peak ion chromatograms corresponding to both the fresh and dry ginkgo leaves. It is apparent from Figure 1a that visually discerning differences between the two complex samples is difficult. However, using the MarkerLynx XS Principal Component Analysis (PCA) function, the grouping pattern of the fresh leaves (A) and the dry leaves (B) becomes clear (Figure 1b). Furthermore, Figure 2a shows the S-plot obtained from the orthogonal partial least-squares-discriminant analysis (OPLS-DA) of fresh and dry leaves. The exact mass retention time (EMRT) pairs on both ends of the S-plot are identified as the leading markers providing the most significant contribution to the grouping.
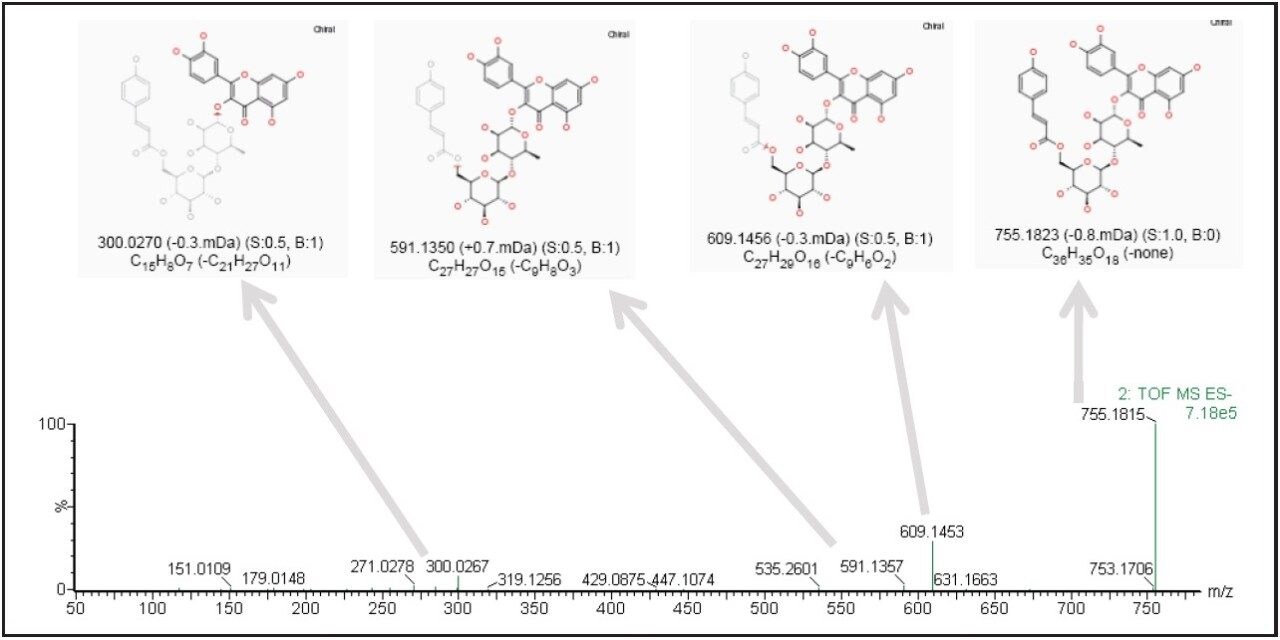
The precursor ion data corresponding to the identified markers can be searched against a database to assist in identification. Furthermore, the associated fragment ion data can be used for structural elucidation to support the proposed component identity. Figure 2b shows a trending plot of one leading marker (m/z of 755.1824), with a possible structure obtained from the ChemSpider database. Figure 3 shows the fragment ion data associated with the same marker; structural elucidation and annotation of this data was performed automatically by the MassFragment Software tool, supporting the proposed compound identification.
Non-target ingredient profiling for complex samples is often a challenging task, but can be handled with ease using the UPLC-SYNAPT G2 HDMS/MarkerLynx XS System.
The combination of UPLC separations and SYNAPT G2’s high quality, exact mass data processed with MarkerLynx XS Application Manager allowed leading markers that differentiate complex sample groups to be confidently identified. Furthermore, the system automates advanced statistical analysis such as PCA and OPLS-DA to complete the profiling workflow.
720003489, May 2010