For research use only. Not for use in diagnostic procedures.
This reference guide provides recommended step-by-step instructions for sample preparation followed by HILIC chromatography of human plasma/serum samples for semi-quantitative lipid analysis.
The purpose of this analysis document is to provide Waters recommended step-by-step instructions for sample preparation followed by HILIC chromatography of human plasma/serum samples for semi-quantitative large cohort lipid analysis. This document includes: details of sample preparation/extraction using IPA, mobile phase buffer preparation, and UPLC-MS/MS analysis of the samples.
This document contains Waters recommendations for:
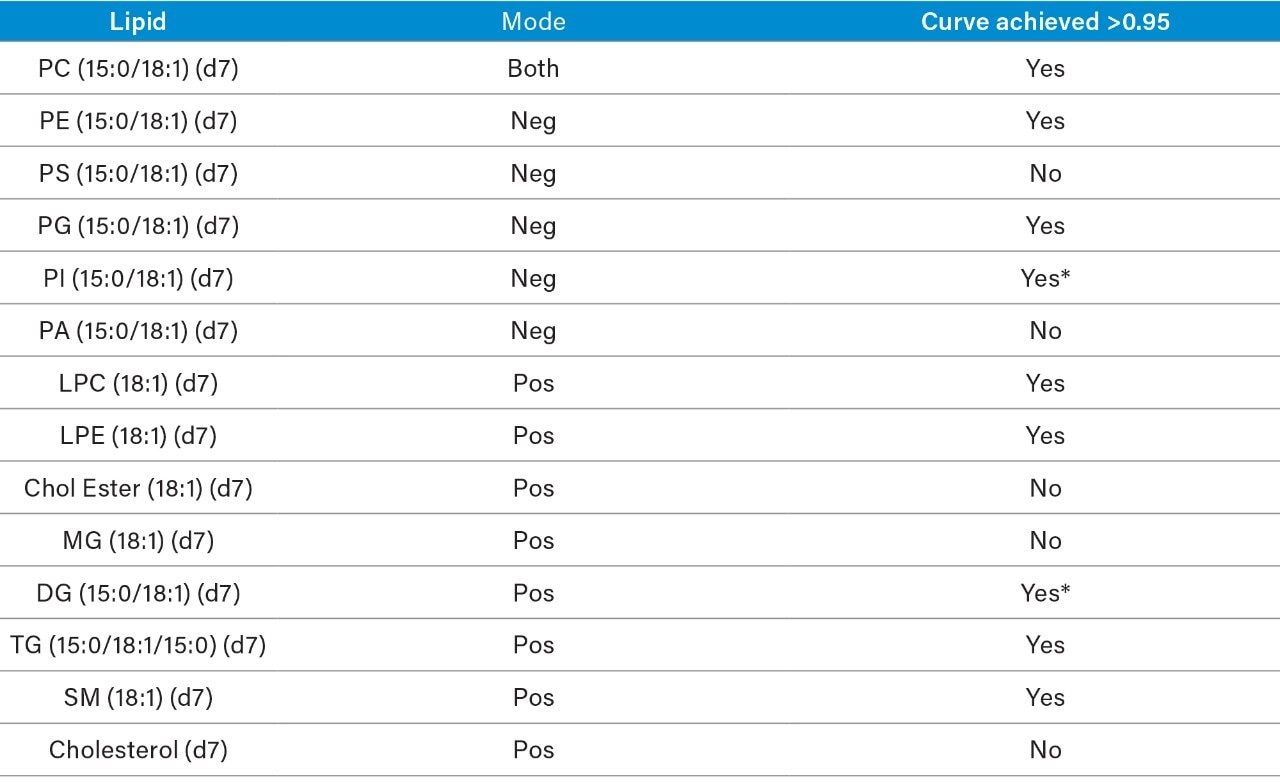
The following method conditions are suitable for the semi-quantification of (polar lipids) phospholipids and ceramides. Non-polar lipids (MG, DG, TG, Cholesterol, Cholesterol Esters, etc.) elute in the solvent front using these conditions and therefore concentration values obtained are for monitoring only and should not be deemed quantitative with this method. Although Free Fatty Acids (negative mode) elute close to the solvent front it is still possible to semi-quantify this class using these conditions.
The LipidQuan MS methods, LC methods, and TargetLynx processing method packages are available for download from the Waters website at www.waters.com/TargetedOmics.
A project batch should not consist of more than 1500 biological samples. Calibration curves will be run at the beginning and end of each batch. If the cohorts are large, then it is advisable to run calibration curves at points throughout the analysis to ensure bracketed data is available that will allow to adjust any drift within the run or to flag any analysis failure.
Prior to sample analysis, blanks and quality controls (QC) injections are performed to demonstrate that the instrument is stable and that results are consistent prior to running valuable biological samples. Ensuring that the instrument is stable prior to sample analysis will also help to maintain consistency between the runs.
In addition to running blanks and QCs prior to the start of analytical runs, QC samples are acquired throughout the analytical run; these intra-run QCs will assist with determining analysis stability and data suitability. These frequent injections allow for instrument changes to be monitored. They can also be used to indicate injection suitability prior to, or post, any instrument issues that result in an analysis stoppage.1
|
CAN |
Acetonitrile |
|
Cer |
Ceramide |
|
CE |
Cholesteryl Ester |
|
CL |
Cardiolipin |
|
DG |
Diacylglycerol |
|
DAG |
Diacylglycerol |
|
FA |
Fatty acyls |
|
FFA |
Free Fatty Acid |
|
GLs |
Glycerolipids |
|
GPs |
Glycerophospholipids |
|
IPA |
Isopropanol (2-Propanol) |
|
LPC |
Lysophosphatidylcholine |
|
MG |
Monoacylglycerols |
|
PA |
Phosphatidic acid |
|
PC |
Phosphatidylcholine |
|
PE |
Phosphatidylethanolamine |
|
PG |
Phosphatidylglycerols |
|
PI |
Phosphatidylinositols |
|
PLs |
Phospholipids |
|
PS |
Phosphatidylserine |
|
QC |
Quality Control |
|
RSD |
Relative standard deviation |
|
RT |
Retention time |
|
SIL |
Stable isotope labelled |
|
SP |
Sphingolipids |
|
SM |
Sphingomyelin |
|
TG |
Triacylglycerol |
|
TAG |
Triacylglycerol |
These volumes can be scaled up or down, ensure that whenever preparing the calibration curve and QCs that the total volume v/v of SPLASH LIPIDOMIX is no more than 5% at any time.
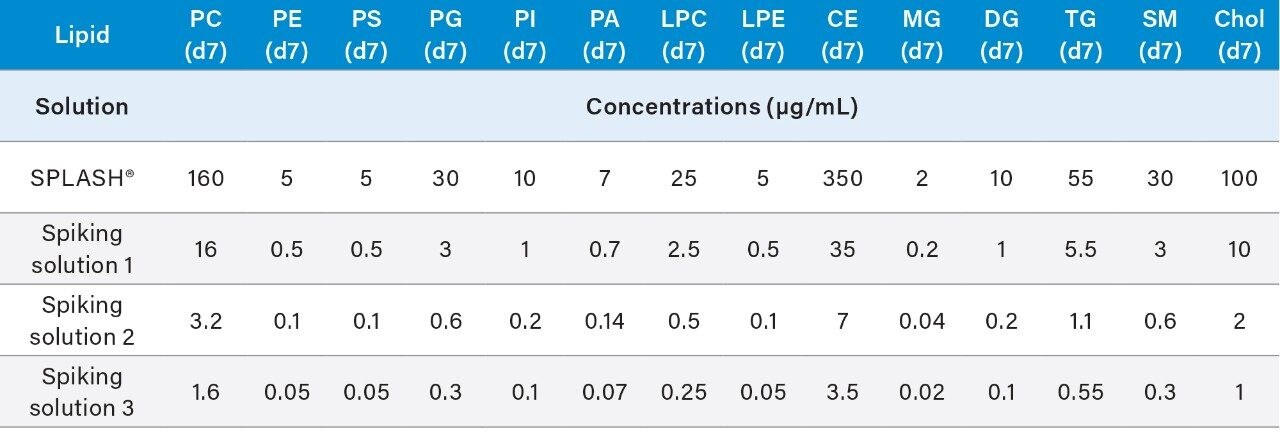
6. Vortex mix each solution for 15 seconds to ensure homogeneity.
7. The spiking solutions may be stored at -20°C (±5°C) for up to 6 months.
1. Transfer 20 μL of spiking solution 2 (Table 1) into suitable glass HPLC vials.
2. Dilute with 480 μL of IPA (chilled to 4–8 °C).
3. Vortex mix for 15 seconds to ensure the solution is homogenous.
4. System blank should be a vial of IPA (chilled to 4–8 °C).
5. This must be prepared as described in section 6.5.
A pool composed of an aliquot from every sample to be analyzed should be prepared. To calculate the amount of sample to remove from each sample for the pool, divide the number of samples by the total pool required (#samples/pool = µL/sample to be removed). The pooled samples will be representative of all the samples in the study and will be combined with the standards mix to form the QCs and the calibration curve standards. Pooled plasma can be aliquoted to microcentrifuge tubes (6.5.2) and stored at -80 °C a day prior to the analysis. At least one aliquot of blank matrix should be prepared from the pooled samples; the blank matrix includes the QCs but not the calibrants (Table 2).
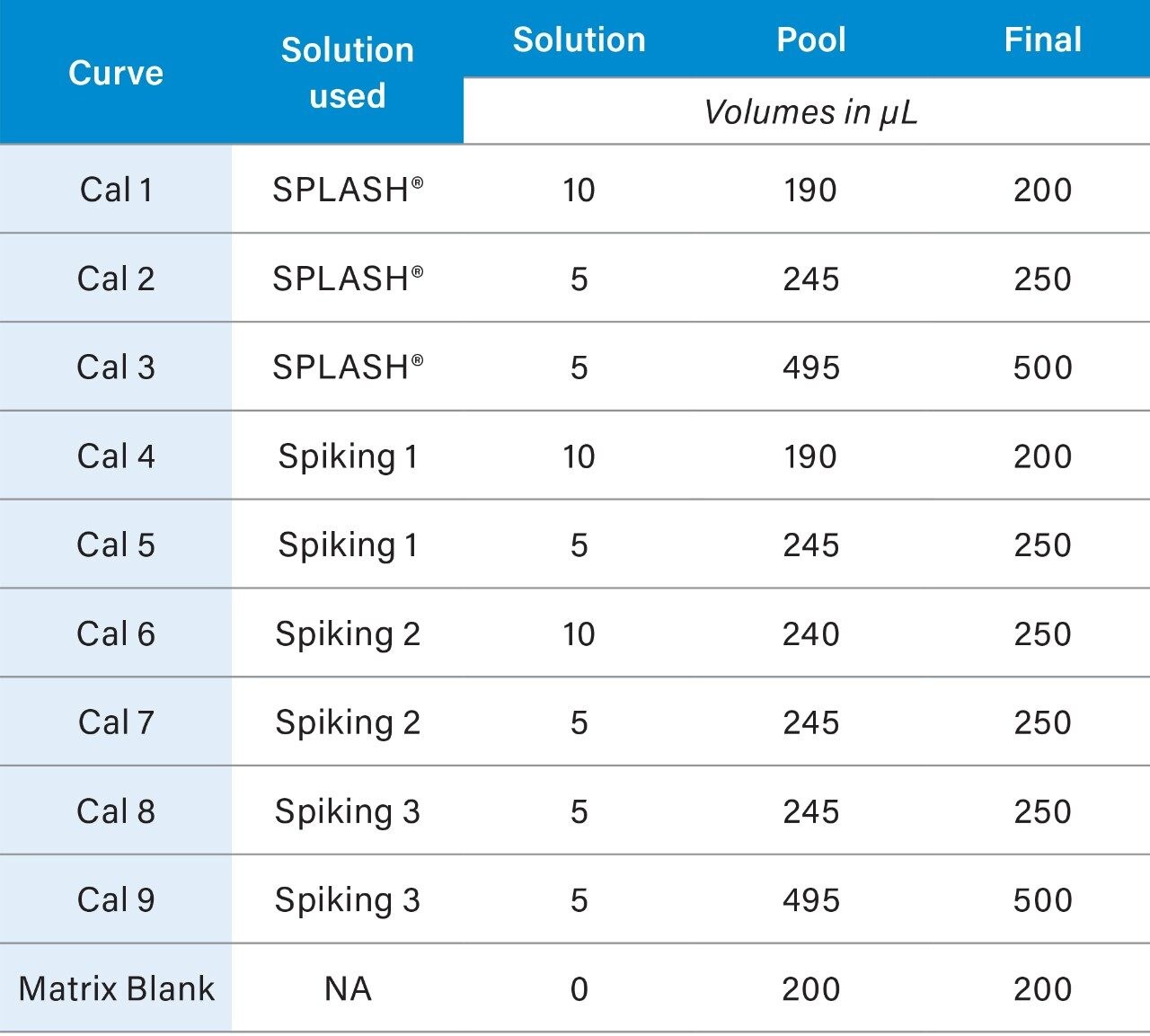
3. The resulting concentrations for the curve are listed below in Table 3.
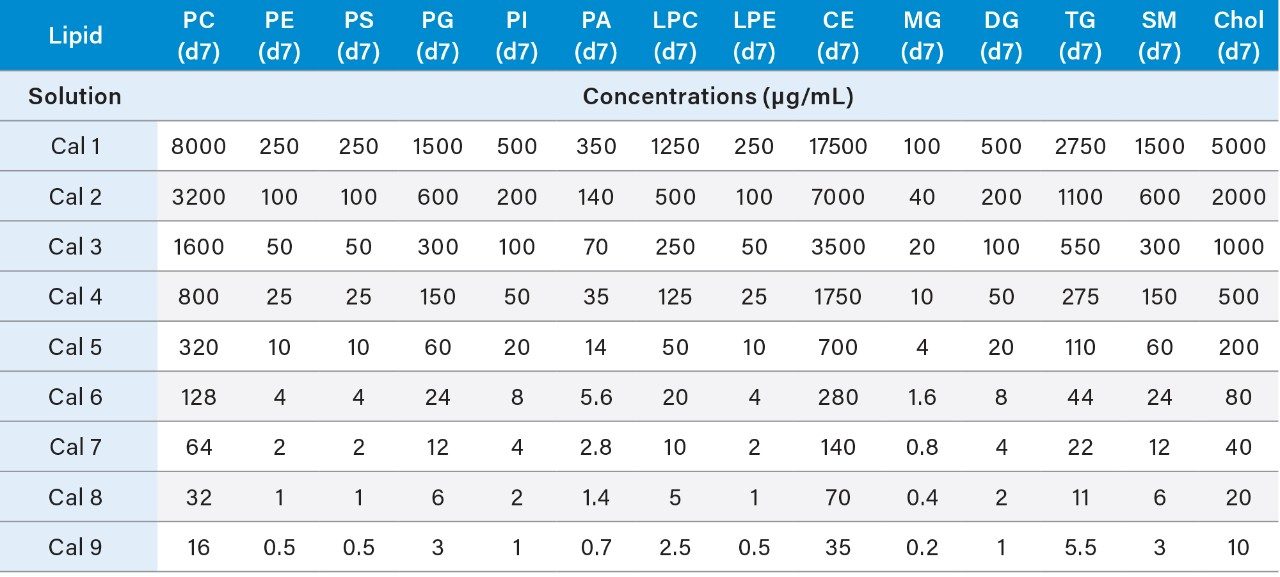
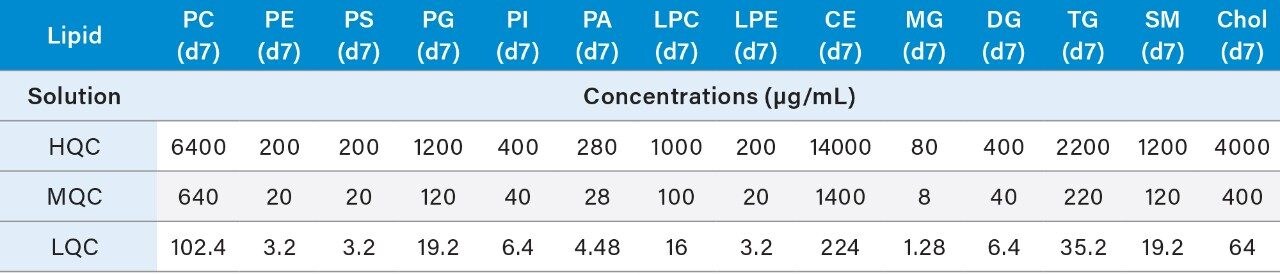
4. Prepare the QC samples using the standards solutions mix prepared in 6.1 and the pooled sample volumes as described below in Table 4. High (HQC), Middle (MQC), and Low (LQC) QC samples should be prepared at 80%, 8%, and 1.275% of the highest concentration of the calibration curve.

5. The resulting concentrations for the curve are listed below in Table 5.
A simple protein precipitation sample preparation procedure should be used with pre-cooled isopropanol (IPA).2
95:5 Acetonitrile:Water, 10 mM Ammonium acetate
50:50 Acetonitrile:Water, 10 mM Ammonium acetate
Scale up as necessary. It is recommended to prepare ALL required analysis mobile phases in one large batch and aliquot into suitable quantities. It is best practice to use a single batch of all solvents and additives during the entire study. This will minimize batch differences seen through the analysis.
9.3.1 Seal Wash
10% acetonitrile in water
1. Measure 900 mL of water.
2. Top up bottle with 100 mL with acetonitrile.
9.3.2 Weak wash (FL)
95:5 (v/v) Acetonitrile:Water
1. Measure 50 mL of water.
2. Top up bottle with 950 mL with acetonitrile.
9.3.3 Strong wash (FL), Needle Wash and Purge (FTN)
100% isopropanol



The LipidQuan MS and LC (contained in the LipidQuan Quanpedia file) settings as well as the TargetLynx processing files are available for download from the Waters website at www.waters.com/targetedOmics.
1. Suggested analysis running order*:
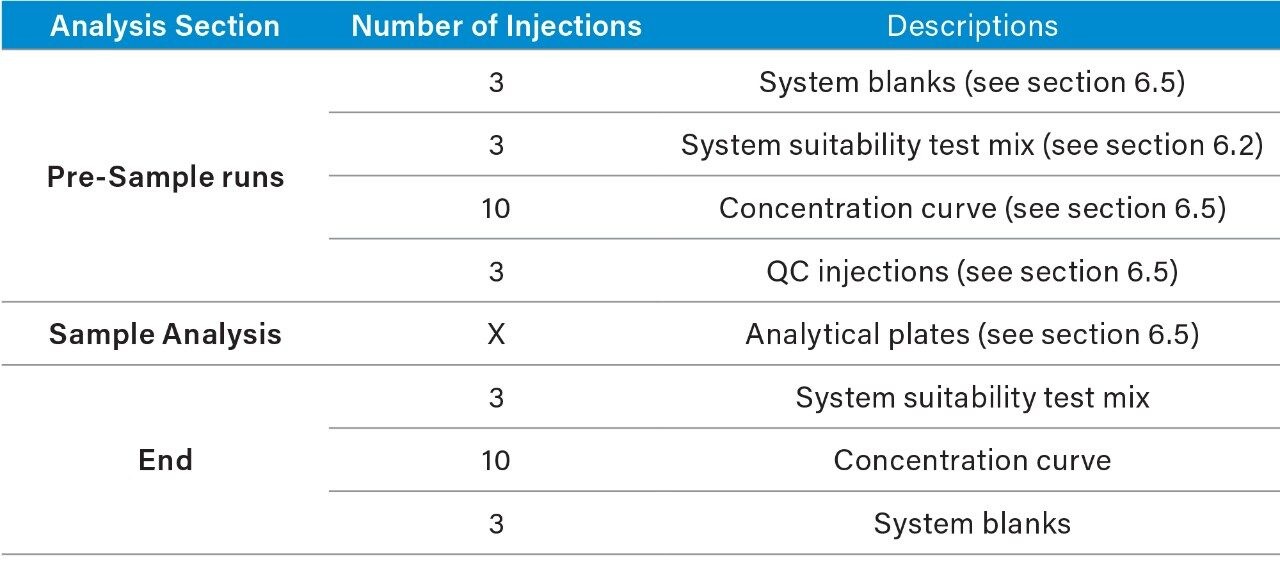
2. System blanks and suitability test mix should be checked prior to continued acquisition. In order to ensure the system is functioning as expected.
3. Should the analysis halt for any reason and the analysis session requires a restart, the analyst should assess the previous analysis. The subsequent course of action will be down to operator discretion.
—Should the issue have impacted multiple injections it may be necessary to re-prepare the affected plate and re-analyze.
—If the issue has not impacted previously injected samples then it is possible to re-start the analysis continuing sample analysis from the last injection. Once the system is operational, the operator must inject a new concentration curve to bracket the new analysis set. The initial set may need to be processed with only a single calibration curve.
720006541, July 2019