
The presence of an increasingly complex array of pharmaceuticals and personal care products (PPCPs) in water bodies throughout the world is placing a greater demand on techniques used to screen for these compounds. In combination with UNIFI, an integrated scientific information system, it is now possible to screen for the presence of PPCPs, their fragments, adducts, and potential metabolites in a routine laboratory environment. This application note describes the use of Waters Screening Platform Solution in combination with the UNIFI Toxicology library to screen a local well water sample for the presence of a large number (>1000) of PPCPs, pesticides, and drugs of abuse.
The presence of an increasingly complex array of pharmaceuticals and personal care products (PPCPs)1 in water bodies throughout the world is placing a greater demand on techniques used to screen for these compounds. Figure 1 illustrates an example of the range of PPCP compounds and classes.
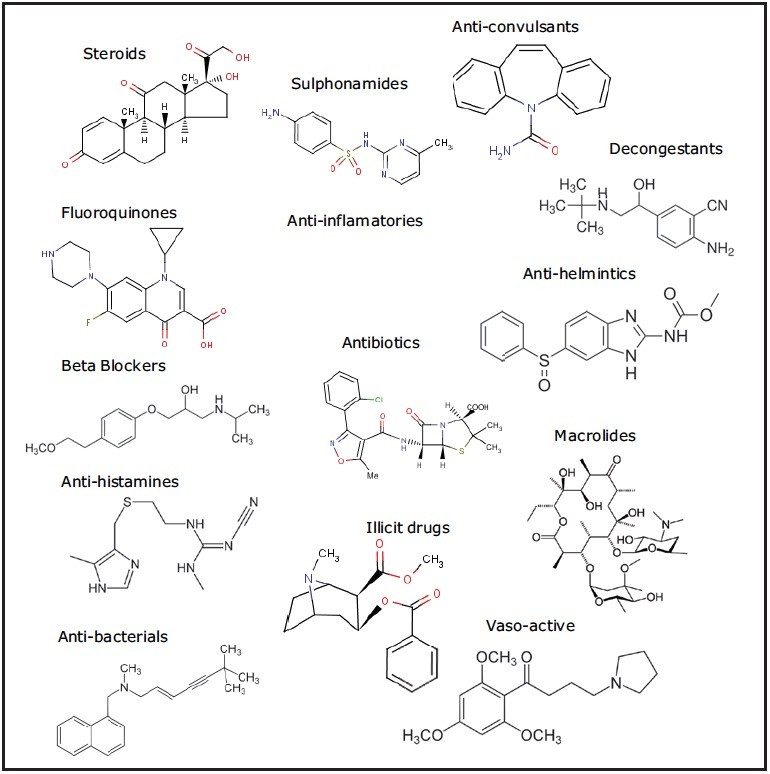
Conventional MS/MS screening methods, utilizing a tandem mass spectrometer, have target list limitations as the maximum duty cycle of the instrument is achieved. Data obtained from a non-targeted acquisition on a high resolution mass spectrometer can be used to target a theoretical unlimited number of compounds. Moreover, information rich datasets collected using UPLC-MSE can be used to reduce the large number of false detects that arise when targeting a large number of compounds with accurate mass as the sole point of contaminant identification. MSE provides accurate mass measurements for both precursor and fragment ion information in a single experiment.
In combination with UNIFI, an integrated scientific information system, it is now possible to screen for the presence of PPCPs, their fragments, adducts, and potential metabolites in a routine laboratory environment. This application note describes the use of Waters Screening Platform Solution in combination with the UNIFI Toxicology library to screen a local well water sample for the presence of a large number (>1000) of PPCPs, pesticides, and drugs of abuse.
The following samples were prepared.
PPCP standards: High level standards (10 μg/L) in UHPLC-grade water
Extracted blank: Treated as a reference. A UHPLC-grade water sample (Fisher Optima), enriched using the protocol in Figure 2.
Extracted well water sample: Treated as the unknown. A well water sample enriched as above.
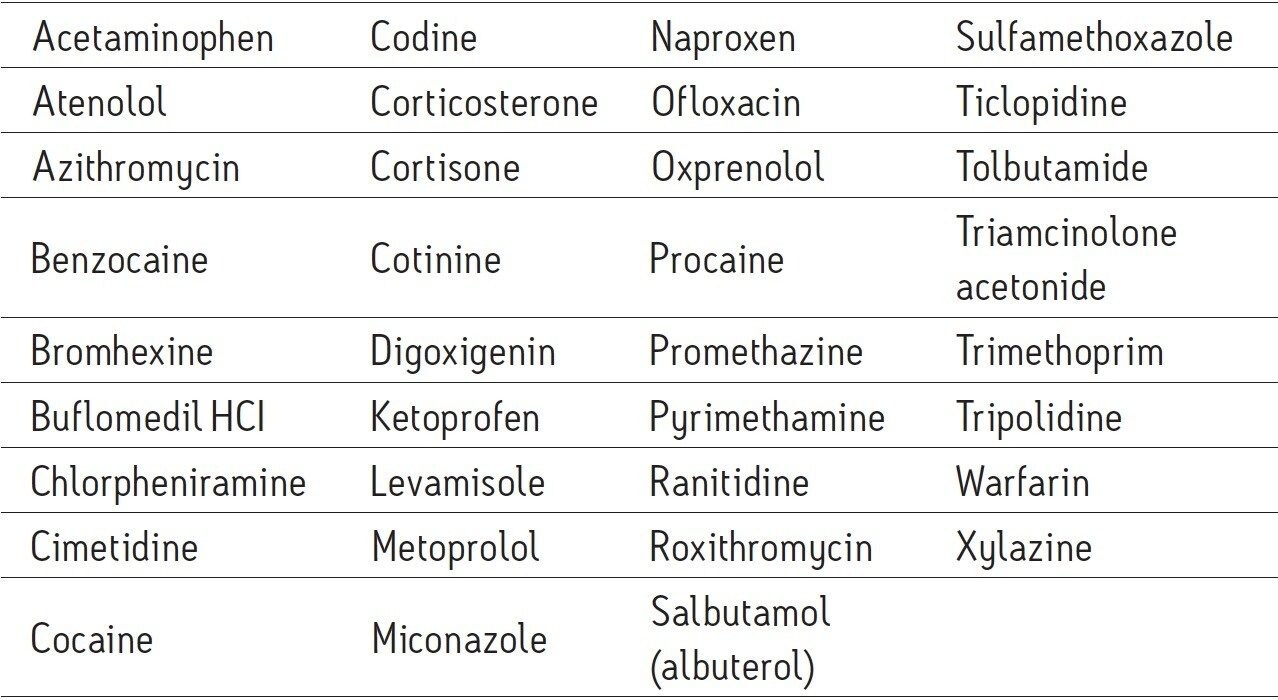
Extracted well water sample, post spike: Enriched as above and post spiked with the 35 PPCPs in Table 1 at a level of 1 μg/L.
Extracted well water sample, pre spike: Well water sample pre-spiked with the 35 PPCPs described in Table 1 (at 1 ng/L) then enriched as above. Pre-spike samples were prepared in duplicate.
Non-extracted well water: Well water sample neither enriched nor spiked.
Extracted calibration standards: Eight levels (1.0, 2.0, 2.5, 5.0, 10.0, 25.0, and 50.0 ng/L) of the 35 PPCPs in Table 1 spiked into the UHPLC-grade water and enriched as above.
Comprehensive datasets were collected using UPLC-MSE data acquisition. MSE uses parallel low and elevated collision energy MS acquisition to provide accurate mass precursor and product ion information in a single injection.
|
LC system: |
ACQUITY UPLC I-Class |
|
Runtime: |
15.00 min |
|
Flow rate: |
0.40 mL/min |
|
Injection: |
100.0 μL |
|
Column: |
ACQUITY UPLC HSS C18 1.8 μm, 2.1 x 150 mm |
|
Column temp.: |
50 °C |
|
Mobile phase A: |
Water with 5 mM NH4HCO2 adjusted to pH 3.0 with formic acid |
|
Mobile phase B: |
Acetonitrile with 0.1% (v/v) formic acid |
|
Time |
Flow rate mL/min |
Composition A |
Composition B |
Curve |
|---|---|---|---|---|
|
0.00 |
0.400 |
87.0 |
13.0 |
Initial |
|
0.50 |
0.400 |
87.0 |
13.0 |
6 |
|
10.00 |
0.400 |
50.0 |
50.0 |
6 |
|
10.75 |
0.400 |
5.0 |
95.0 |
6 |
|
12.25 |
0.400 |
5.0 |
95.0 |
6 |
|
12.50 |
0.400 |
87.0 |
13.0 |
6 |
|
15.00 |
0.400 |
87.0 |
13.0 |
6 |
|
MS system: |
Xevo G2-S QTof |
|
Ionization mode: |
ESI+ |
|
Scan time: |
0.2 seconds |
|
Capillary voltage: |
1.0 kV |
|
Sampling cone: |
20.0 V |
|
Source temp.: |
120 °C |
|
Desolvation temp./gas: |
550 °C/1000 L/H |
|
Mass range: |
m/z 50 to 1200 |
|
MSE low energy: |
4.0 V |
|
MSE high energy: |
10.0 to 45.0 V |
|
LockSpray solution: |
Leucine enkephalin |
|
LockSpray mass: |
m/z 556.2766 |
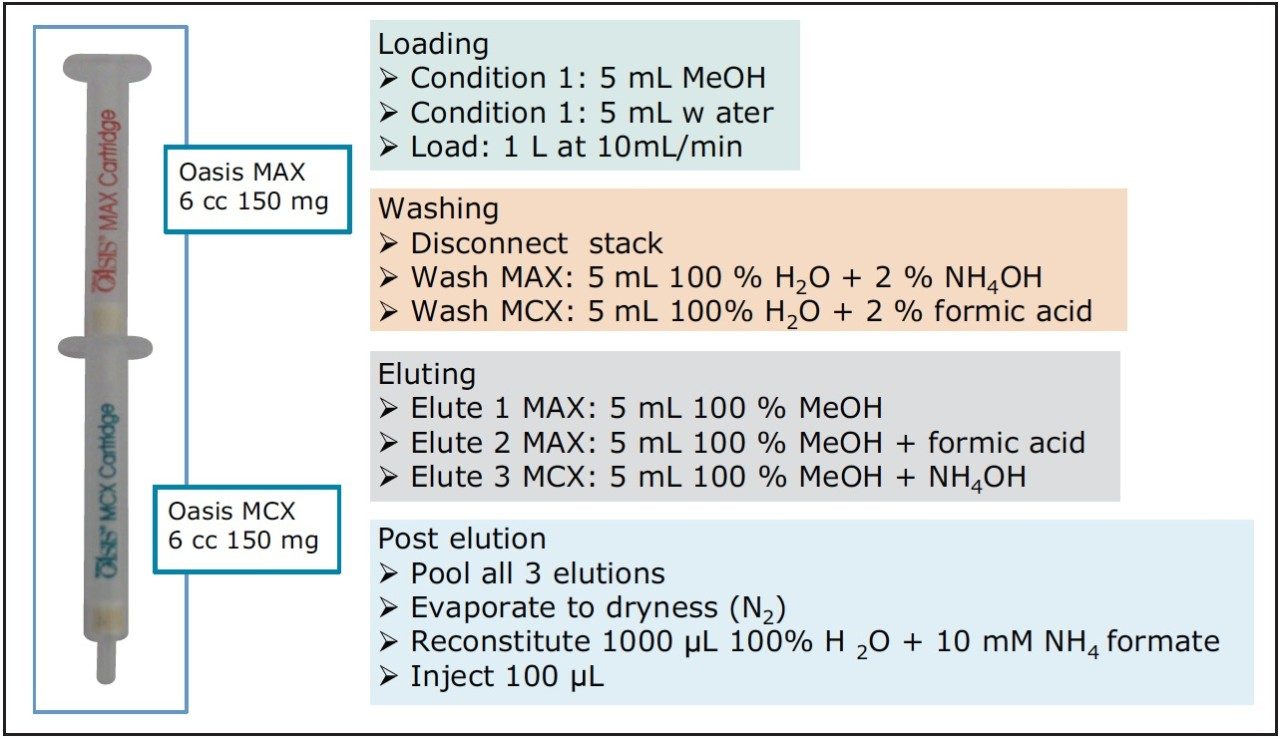
Due to the wide chemical diversity of compounds described as PPCPs, the extraction and separation of the many classes and structures poses a major analytical challenge. A method employing mixed mode solid phase extraction was used to prepare samples, as shown in Figure 2 and described in a previous work.2
The concentration level of the post spiked compounds was chosen to account for the 1000-fold enrichment achieved in the sample preparation. A concentration of 1 ppt (1 ng/L) in the sample equated to an in-vial concentration at the same level as the post spike sample, 1 ppb (1 μg/L).
All MSE data were collected and processed within the UNIFI, Scientific Information System. Data within UNIFI is passed through the apex peak detection and alignment processing algorithms.3 This enables related ion components to be grouped together and analyzed as a single entity. Charged species, salt adducts, and fragments are all automatically aligned and grouped, so that all this information can be used to automatically interpret the data.
The Forensic Toxicology Screening Application Solution with UNIFI comes with pre-defined LC-MS conditions and processing parameters encompassed within an analysis method.4 The Toxicology Library in UNIFI is comprised of over 1000 compounds including many PPCPs, such as drugs of abuse, veterinary medicines, and pharmaceuticals. Library entries contain names, formula, mol files, retention times, and theoretical accurate mass fragment ions.
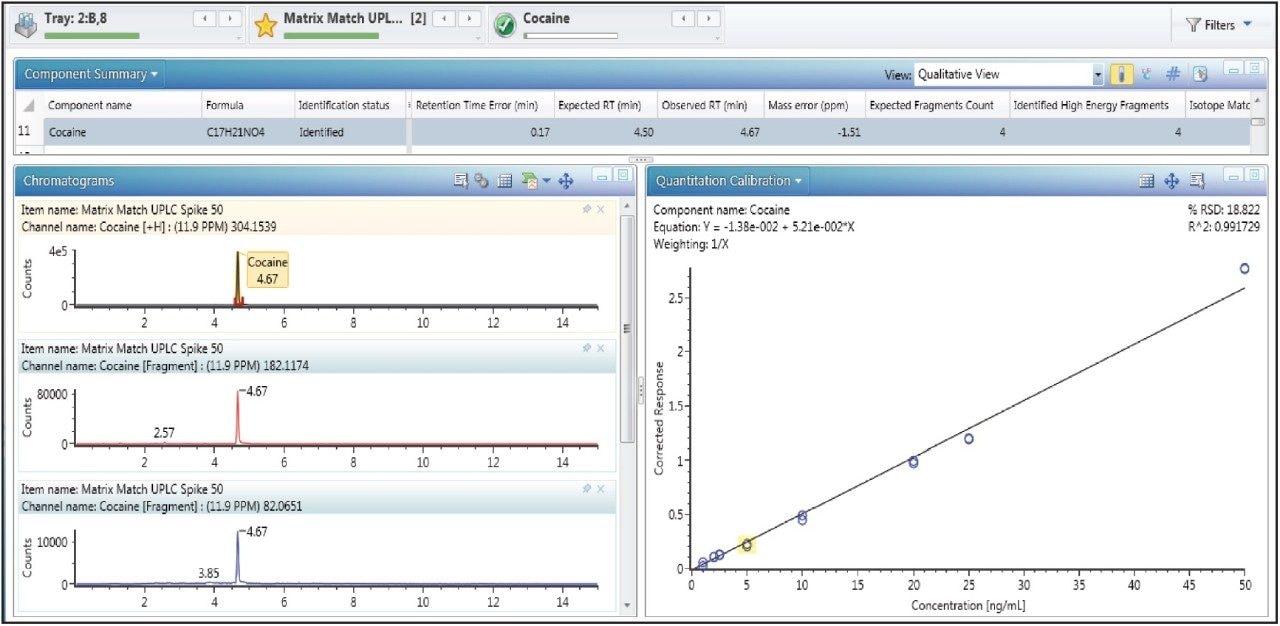
The analysis of compounds spiked into the well water sample demonstrates the applicability of the method for targeted analysis. An example of a calibration curve for one of the standards is shown in Figure 3. Of the 35 standards included, none were detected as incurred residues in the well water sample.
In order to determine whether other compounds were present, the Toxicology Library with up to three adducts (H+, Na+, K+) for each entry was employed. It has previously been demonstrated that screening against a large library using the accurate mass of only one diagnostic ion can result in an unreasonable number of false detects.5 The importance of well resolved, reproducible chromatography in order to reduce the false detect rate has also been highlighted in previous work.6 In the approach used here, full advantage was taken of both retention times and accurate mass fragment ions that are stored in UNIFI’s scientific library. With a mass accuracy tolerance of 5 ppm, a retention time tolerance of 0.5 minutes, and the requirement of at least one fragment ion detection, four incurred residues were found to be present in the well water sample. These are shown in Table 2 and Figure 4.
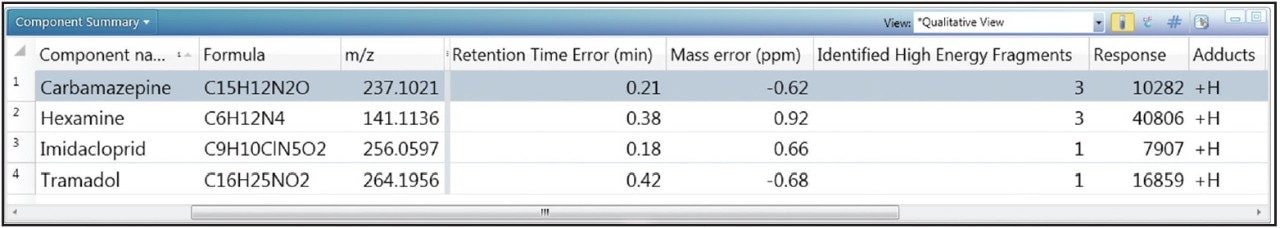
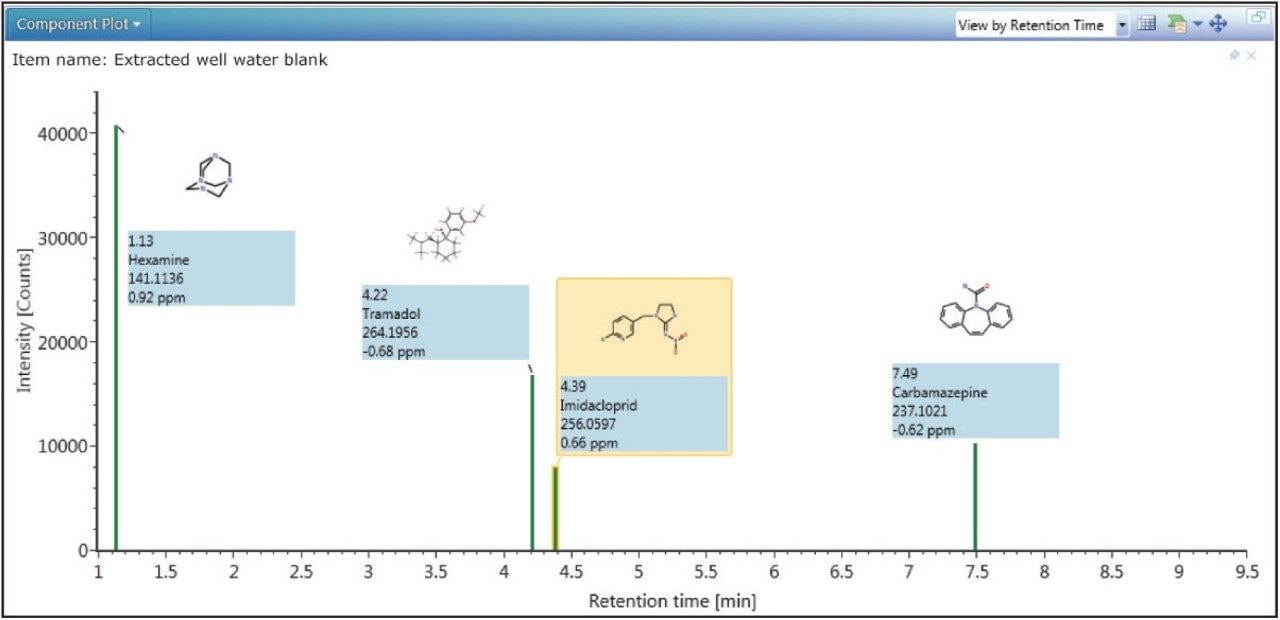
Readily available standards and an existing MS/MS tandem quadrupole method enabled the confirmation and quantification of carbamazepine and imidacloprid. This was achieved using standard addition in a non-extracted well water sample, which was then injected directly for analysis using the ACQUITY UPLC I-Class System with the Xevo TQ-S tandem mass spectrometer (data not shown). Concentrations were found to be 0.31 ng/L and 0.58 ng/L respectively. Tramadol and hexamine were neither confirmed nor quantified.
In UNIFI, the criteria set for identification is completely customizable, giving maximum flexibility to users. The bar chart in Figure 5 shows the effect of changing the retention time tolerances for identification with the sample containing the 35 spiked compounds shown in Table 1. With a mass accuracy tolerances of 5 ppm (SANCO 12495 guidelines) and no retention time or fragment ion criteria, 735 detections were returned (red bars at the far right ofFigure 5). A reduction in false detects was observed when incorporating and decreasing a retention time tolerance, (red bars in Figure 5). Even with a retention time tolerance of 1 minute, the number of detects are reduced by over 65%. However, if this tolerance is set too low, false negatives may arise, as shown for the 35 spiked compounds as shown for the 35 compounds at a tolerance of 0.25 minutes or below (purple bars in Figure 5). A considerable decrease in false detects is observed when fragment ion presence is used to enhance the identification (green bars in Figure 5).
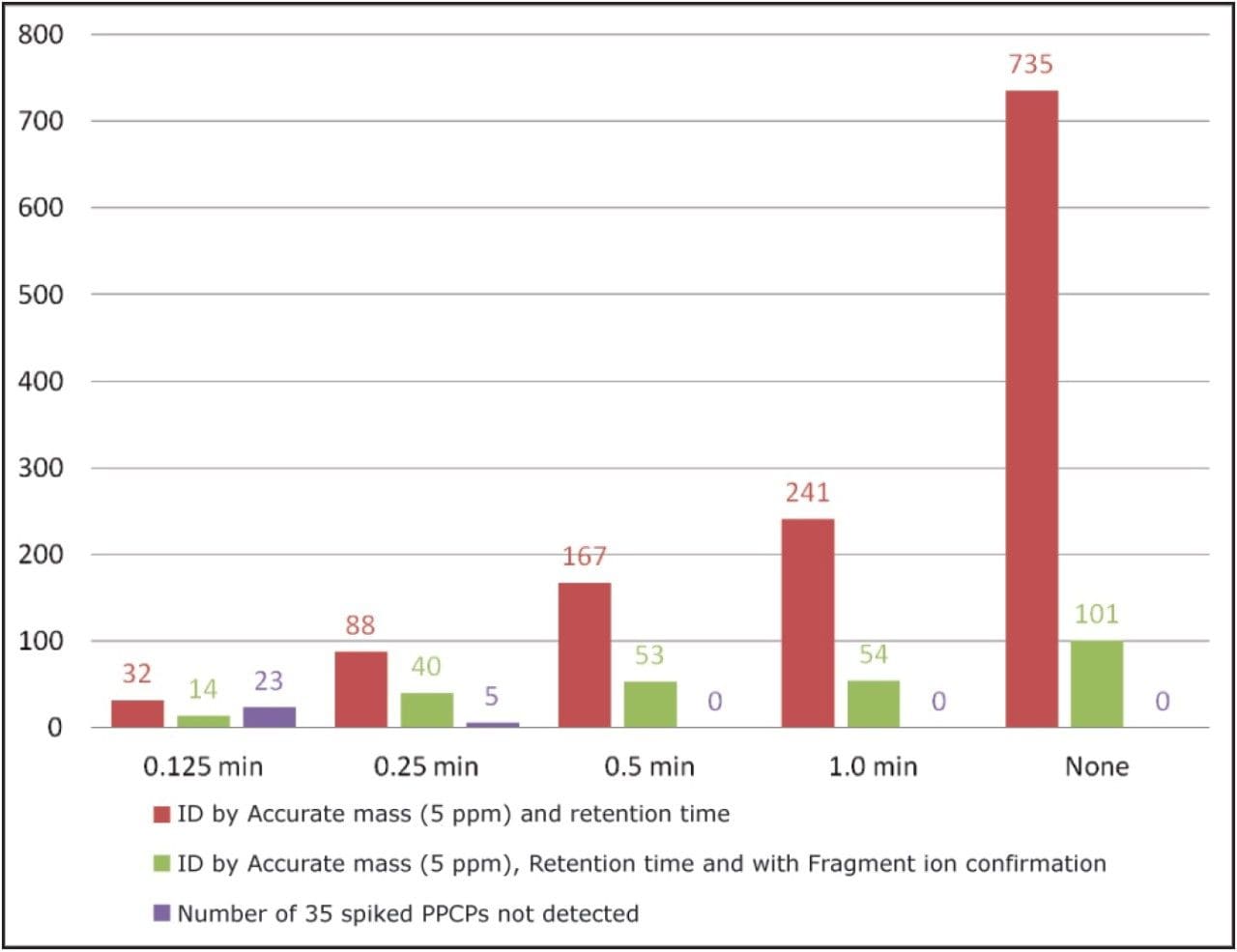
The key to success with HRMS screening is to use wider tolerances for multiple identification criteria. This results in a considerable reduction of false detects while minimizing false negatives. UNIFI provides the flexibility for users to combine criteria in a simplified workflow, enabling rapid data review and improved analyst efficiency.
Four compounds of interest were detected during an HRMS screening of a locally sourced well water sample enriched using an SPE protocol.
720004810, October 2013