The Waters UPLC Amino Acid Analysis Solution provides significantly reduced analysis time of amino acids and baseline resolution of 17 different amino acids found in wine.
Amino acids serve as the nitrogen source for yeasts during the fermentation of wine. Hernández-Orte et al1 have reported that amino acids are also a mark of the quality of wine because they contribute to the wine’s taste, aroma and color. Besides scientifically determining the quality of wine, analysis of amino acids can also classify a wine based on its amino acid concentrations rather than relying solely on the human palate.
Previously, methods for analyzing amino acids in wine have taken anywhere from 46.5 minutes on a Luna C18 bonded silica column (4.6 x 250 mm, 5 μm) protected by a 4.6 x 20 mm sentry guard column1 to 25 minutes on a narrow bore C18 HP Amino Acid Analysis column (2.1 x 200 mm) protected by a 2.1 x 15 mm guard column.2
The Waters UPLC Amino Acid Analysis Solution performs this separation in only 8.5 minutes on an AccQ•Tag Ultra Column (2.1 x 100 mm) with an in-line filter, which is 4.8 and 2.6 times faster, respectively, than previous HPLC methods, while still maintaining baseline resolution of a 17 standard amino acid mixture (Figure 1).

Twenty wines, four of each variety (shiraz, chardonnay, cabernet, sauvignon blanc, and merlot) were purchased from a local liquor store. All samples were stored at room temperature until analysis. Amino acid standard solution, AccQ•Tag Ultra borate buffer, and AccQ•Tag Ultra reagent were all included in the UPLC Amino Acid Analysis Application Solution. Water was purified with a Milli-Q system (Millipore, Billerica, MA).
The Waters ACQUITY UPLC System consisted of a Binary Solvent Manager (BSM), a Sample Manager fitted with a 2 μL loop, and a Tunable UV (TUV) detector. The system was controlled and data collected using Empower 2 Software. Separations were performed on a 2.1 x 100 mm ACQUITY UPLC AccQ•Tag Ultra Column with an in-line filter at a flow rate of 0.70 mL/min.
Column temperature was set at 55 °C and injection volumes for all samples and standards were 1.0 μL. Water/acetonitrile (95:5) was used as the weak needle wash solvent and water/acetonitrile (5:95) was used as the strong needle wash solvent. Mobile phase components and gradient conditions are outlined in Table 1. Detection was set at 260 nm using a sampling rate of 20 points per second and a time constant of 0.4 seconds.
An 80 μL aliquot of AccQ•Tag Ultra borate buffer was added to a total recovery vial. Wine (20 μL, previously diluted 1:8 with water) was added along with 20 μL of reconstituted AccQ•Tag Ultra reagent into the vial. The vial was placed on a vortex mixer for 5 seconds and then allowed to stand at room temperature for 1 minute before being placed in a heating block at 55 °C for 10 minutes. After 10 minutes, vials were removed and analyzed using the ACQUITY UPLC System. All wine samples were prepared and analyzed in triplicate.
A standard solution of 17 amino acids, included as part of the AccQ•Tag Ultra Chemistry Package, was prepared. The standard consisted of a one point calibration curve with standard concentrations of 50 pmoles/μL for all amino acids except cystine, which had a concentration of 25 pmoles/μL (Figure 1).

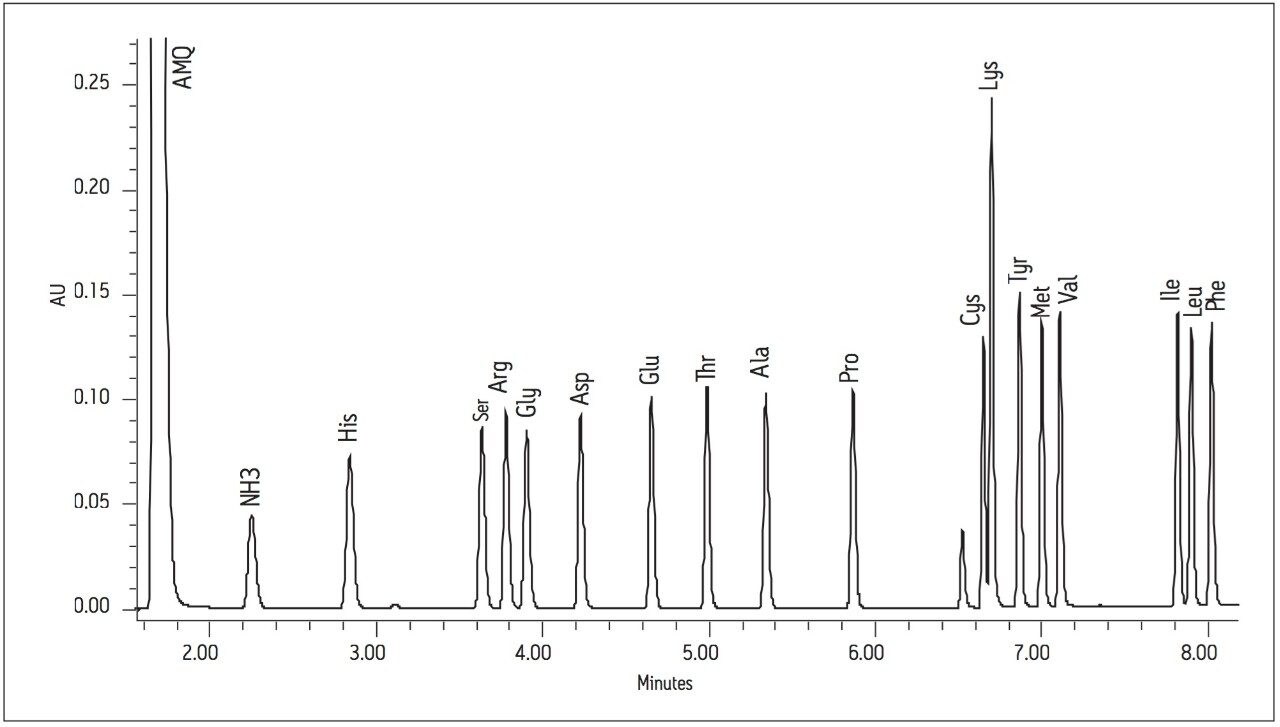
Using the Waters UPLC Amino Acid Analysis Solution, each of the 17 amino acids was baseline resolved in less than 10 minutes. The analysis time of 8.5 minutes was significantly faster than the previous reported times of 45 minutes and 26 minutes. Figures 2 and 3 show the chromatograms of sauvignon blanc 2 and cabernet 4, respectively.
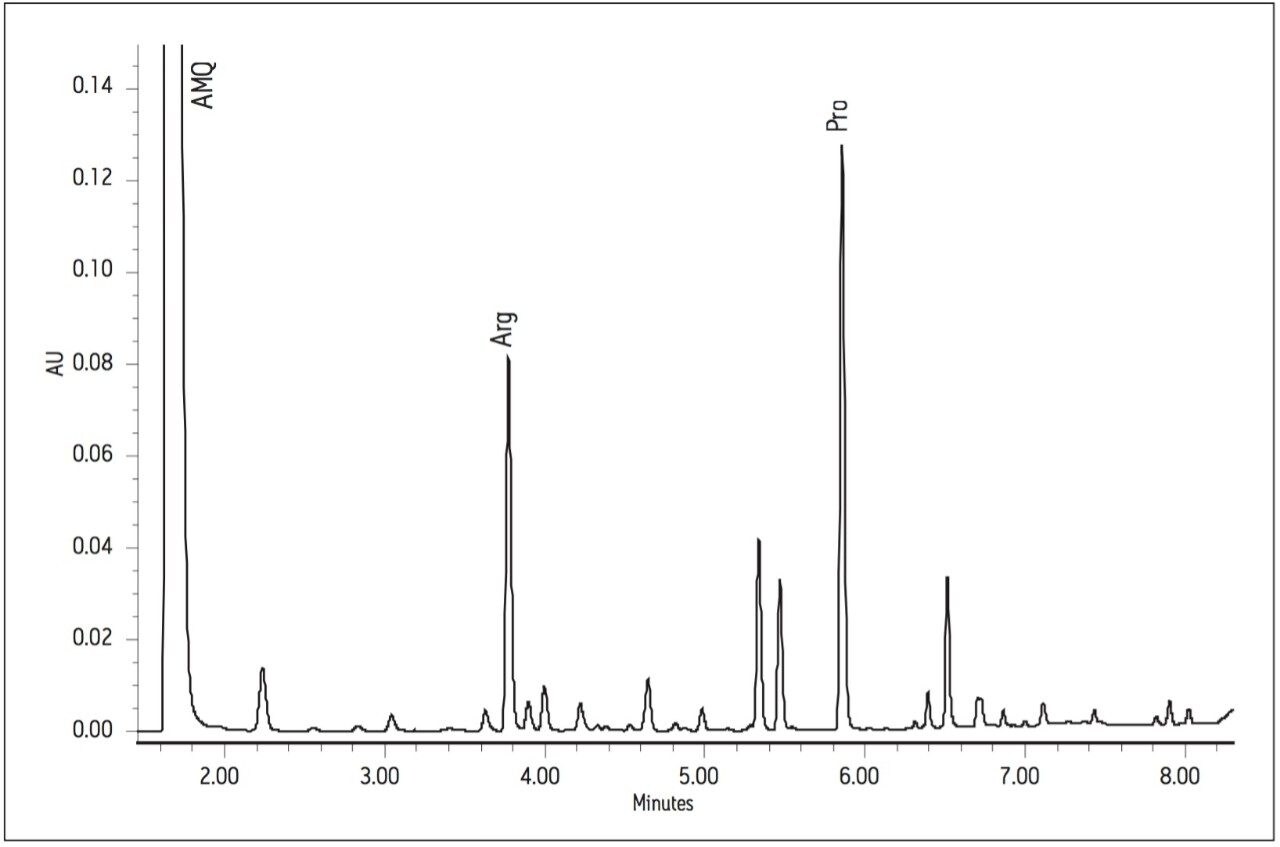
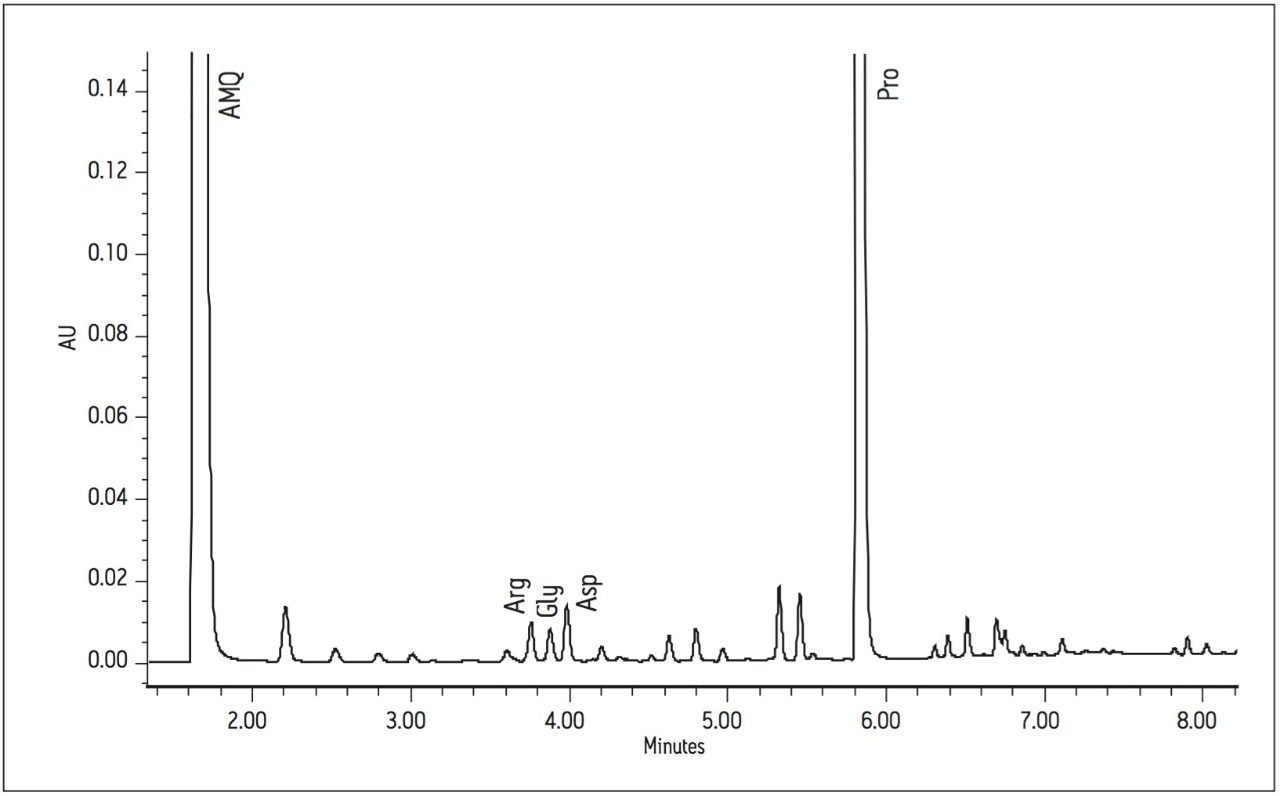
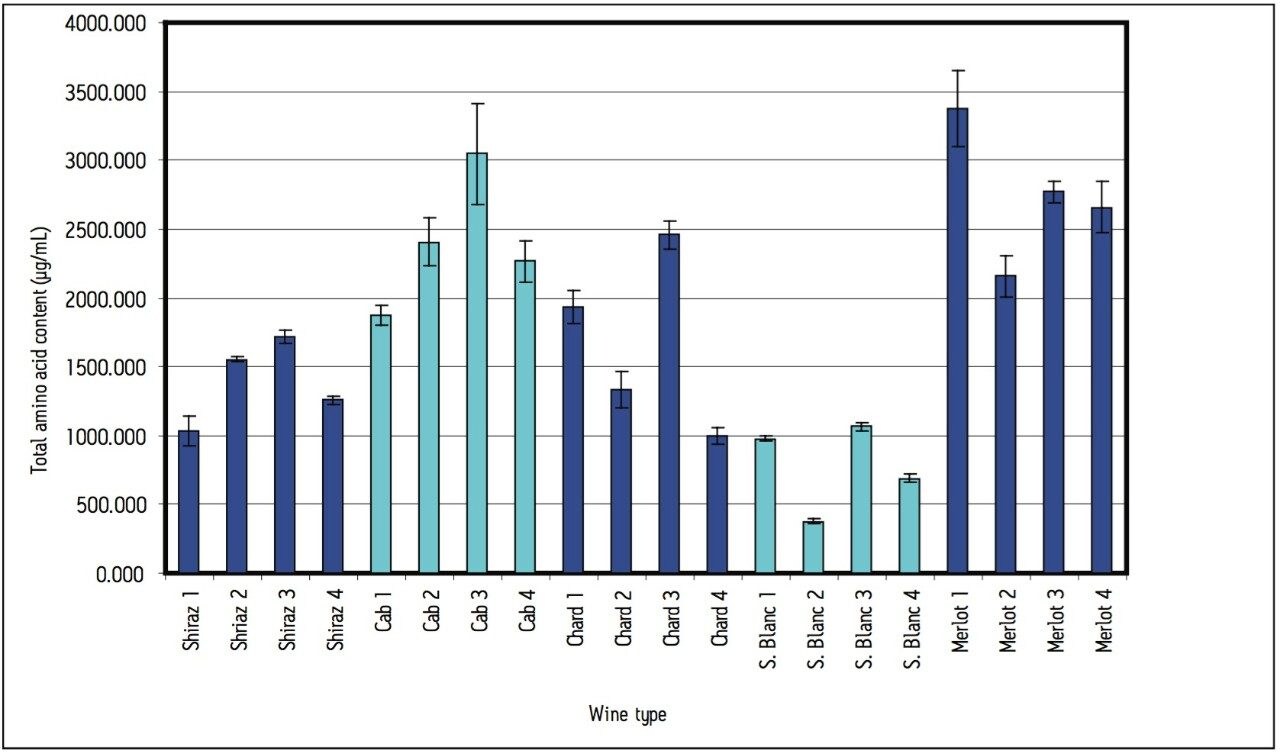
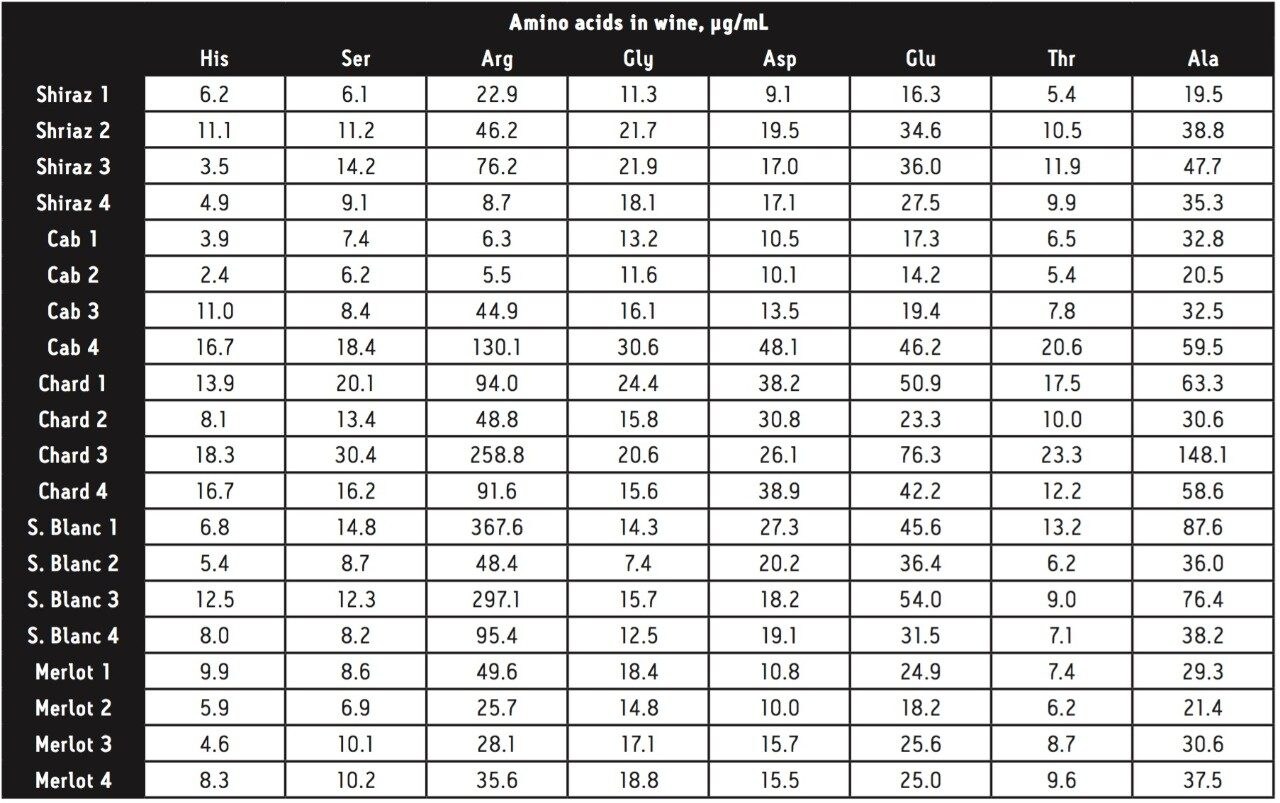
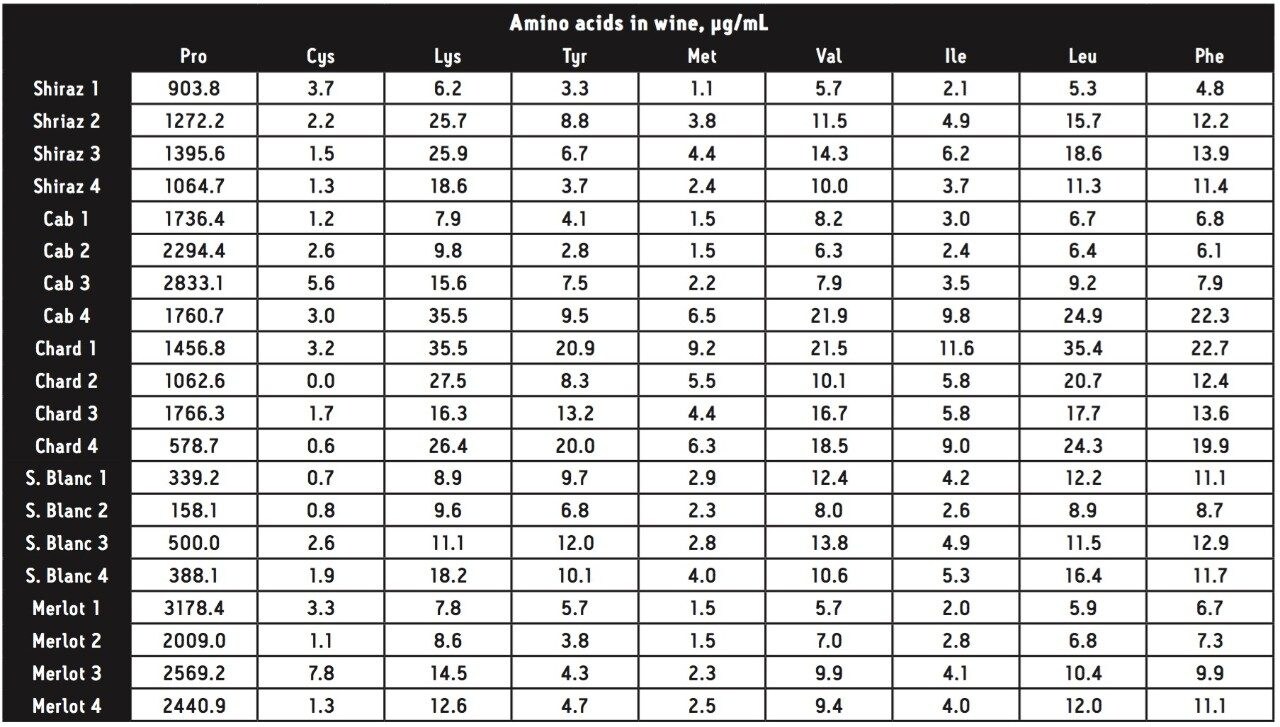
Area counts from injections of derivatized amino acids in the wine samples were compared to the response from the derivatized amino acid standard solution for each amino acid. Concentrations (in μg amino acid per mL) were calculated using Empower 2 Software. All 17 of the amino acids present in the standard solution were found in all of the wine samples, with the exception of cystine, which was not found in one of the chardonnay samples. The total amino acid content (sum of the 17 amino acids in the standard) for each wine can be seen in Figure 4.
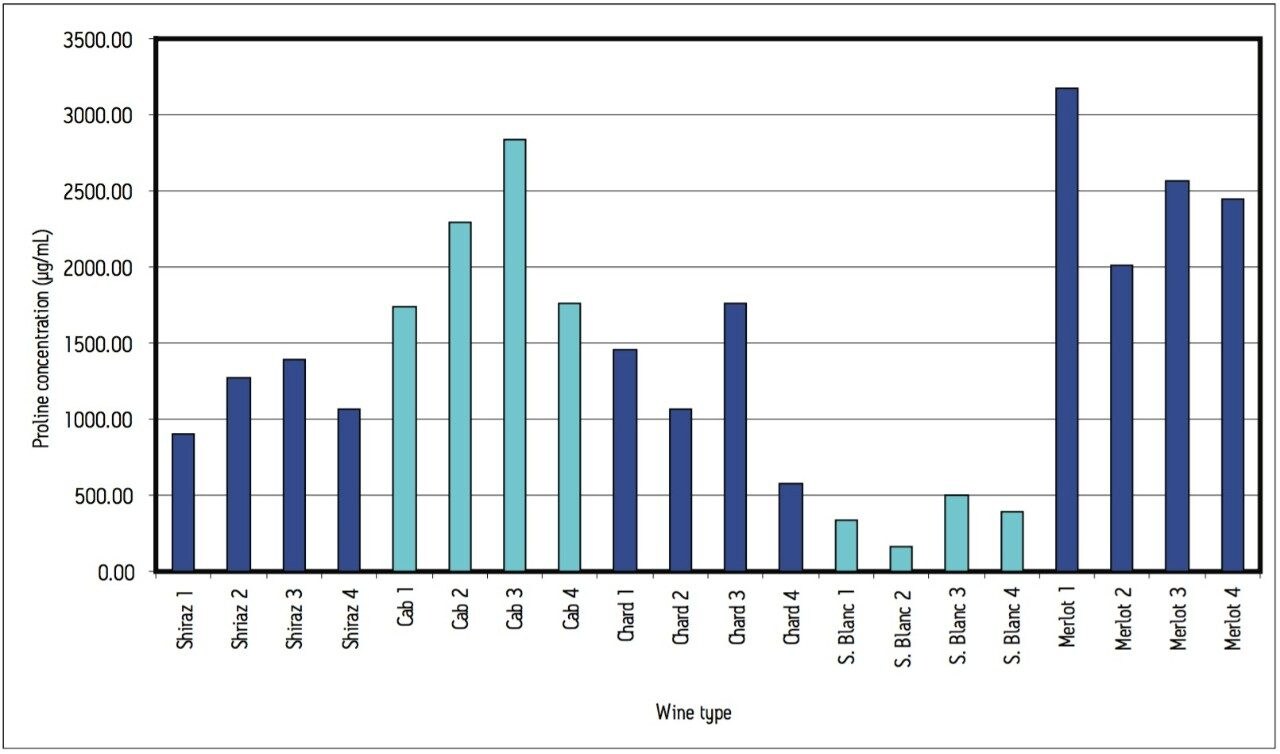
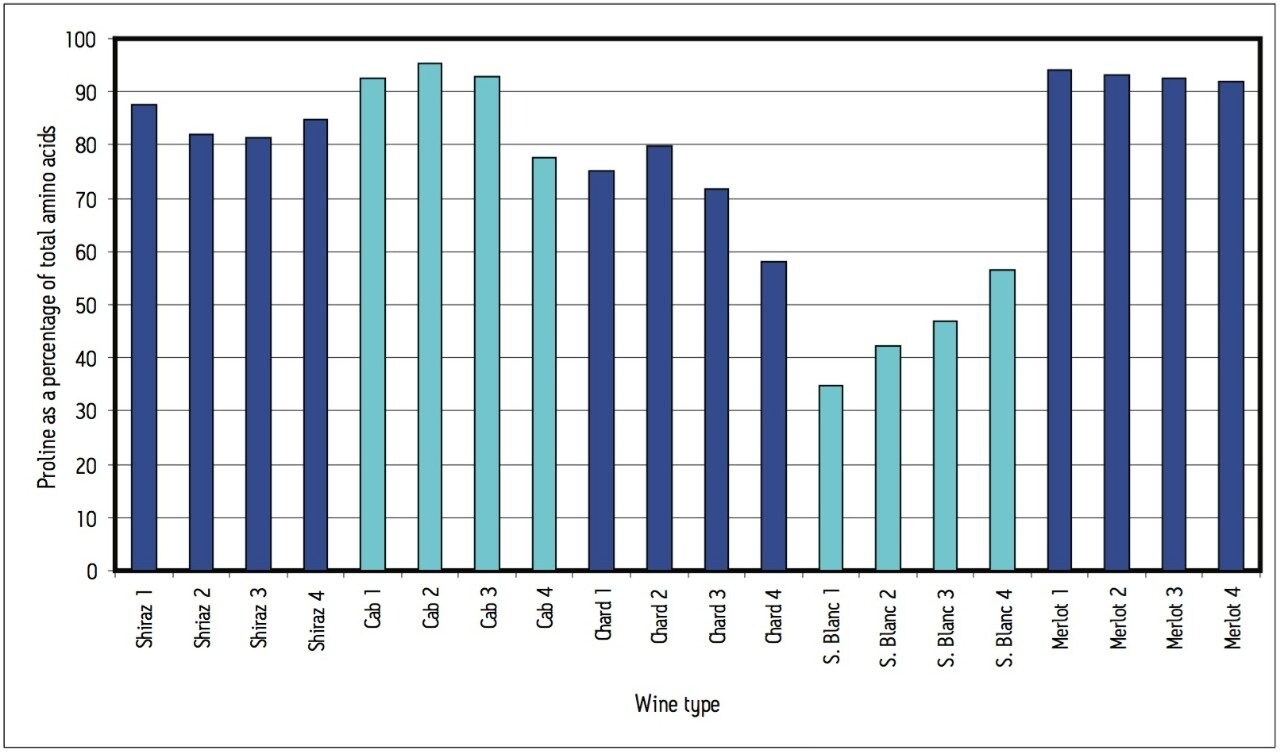
A cursory examination of the data set (Table 2) showed proline to be the most abundant amino acid throughout all 20 different wine samples with the exception of sauvignon blanc 1, which contained slightly more arginine than proline (367 and 339 μg/mL respectively). Concentrations of proline ranged from 158 μg/mL in sauvignon blanc 2 (42% of the total amino acid content for that wine) to 3,178 μg/mL in merlot 1 (94% of the total amino acid content for that wine). Results can be seen in Figures 5 and 6.
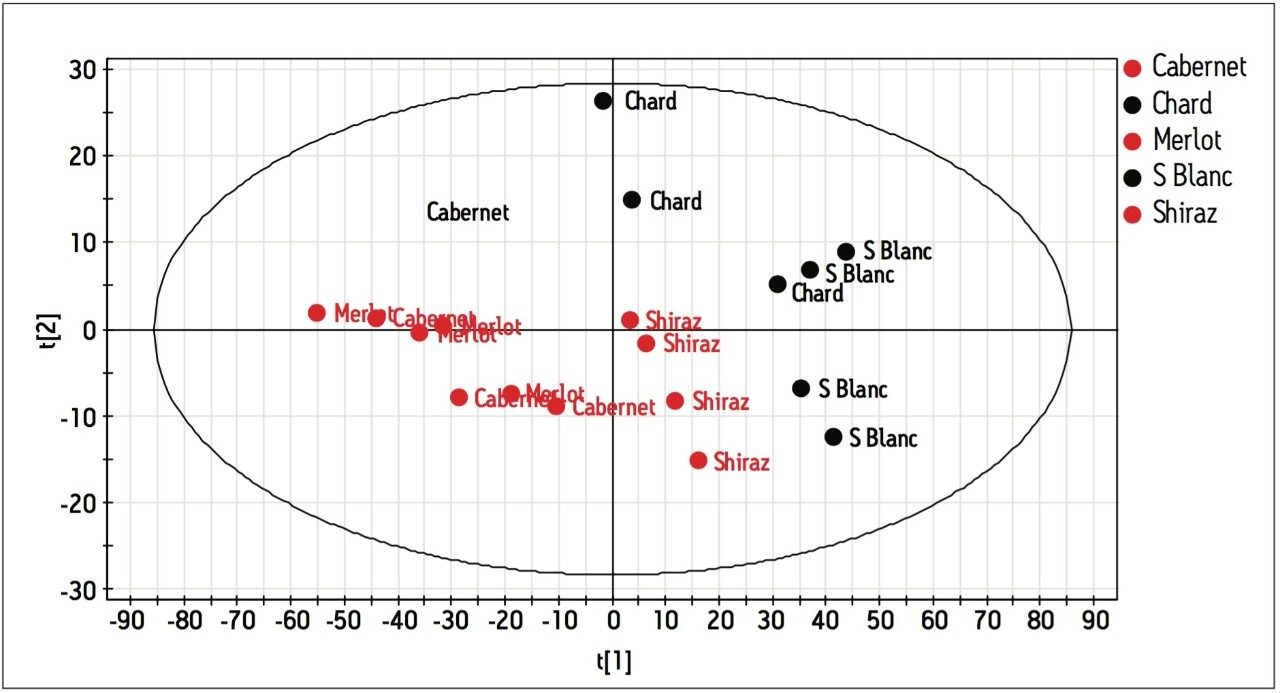
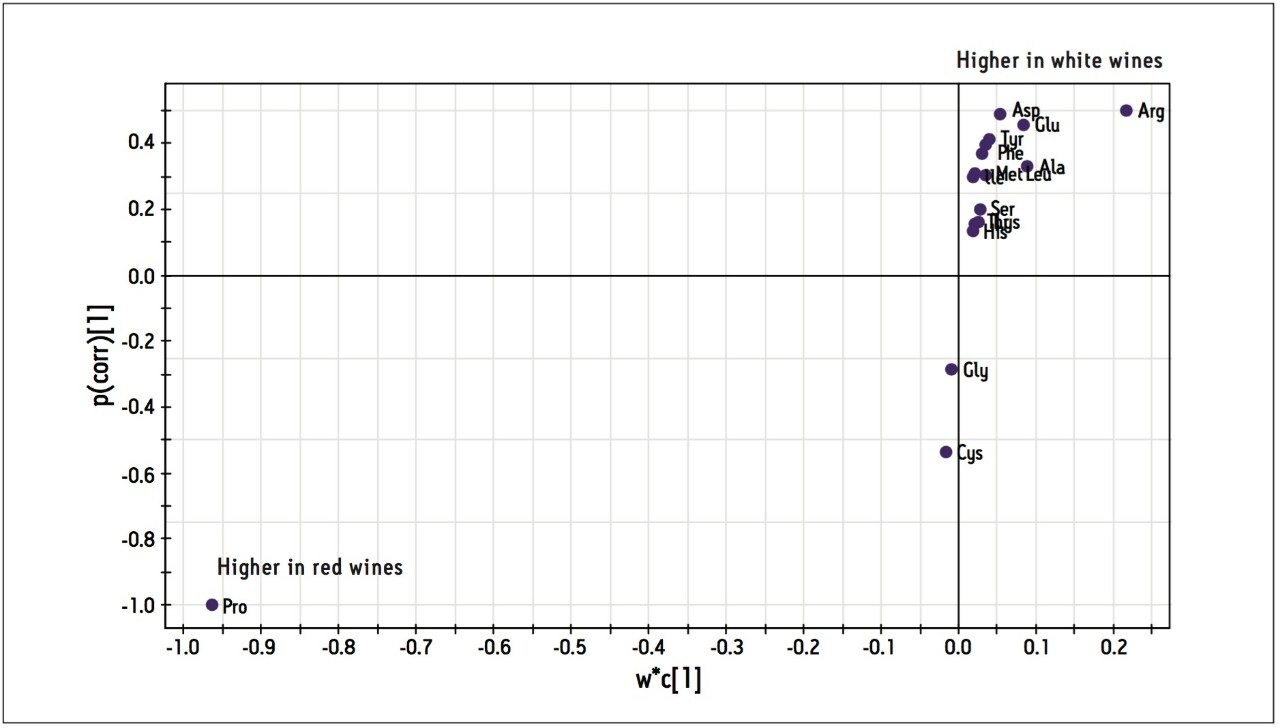
After proline, analysis of Table 2 becomes rather complicated. Further analysis of the data is aided by using multivariate statistical methods (SIMCA-P+ version 11.5 from Umetrics3), i.e. Partial Least Squares Discriminate Analysis (PLS-DA) and Orthogonal Partial Least Squares (OPLS). As shown in Figure 7, PLS-DA clearly shows the variance between the wines, differentiating not just red from white but also the individual varietals. Figure 8 indicates that high proline is the distinguishing feature in red wines while higher arginine, aspartate and glutamate concentrations are indicative of a white wine.
The Waters UPLC Amino Acid Analysis Solution provides significantly reduced analysis time of amino acids and baseline resolution of 17 different amino acids found in wine. The ability to decrease analysis time while also providing baseline resolution results in more productive and efficient laboratories. Applying the Waters UPLC Amino Acid Analysis Solution will greatly benefit any laboratory interested in analyzing amino acids.
Further data analysis using more advanced tools, such as Principal Component Analysis (PCA) and Orthogonal Partial Least Squares (OPLS), provides scientists with additional information from the large, complex data sets in a simplified format.
According to Herbert et al,2 “In order to have routine analysis for wine certification” an amino acid method for wines must be able to accurately detect and quantitate the most prominent amino acids in wine, must have low detection limits for other amino acids and must have a short analysis time. The Waters UPLC Amino Acid Analysis Solution meets these demanding requirements and could be used for wine certification, quality control or identification of adulteration.
720002044, February 2007