
Multi-analyte screening methodologies are essential for monitoring food and environmental samples across the globe. The goal of these methods is to eliminate the compliant samples and to identify the non-compliant samples for subsequent confirmation and quantification. Sensitivity must be in line with the relevant regulatory limits for residues in complex matrices. Also, a method must be validated in accordance with legislative requirements. This method would ideally be rapid, cost effective and a streamlined process, from sample preparation to reporting results.
Many laboratories are progressing towards high-resolution mass spectrometry (HRMS) screening techniques that, in theory, can monitor for an unlimited number of targets at the same time as providing information to help discover unknown compounds or metabolites of interest.
The ease of use and efficacy of a non-targeted, data independent, analysis type (MSE and HDMSE), coupled with a state-of-the-art scientific information system (UNIFI) for multi-analyte screening in food and environmental samples is demonstrated with this case study involving an authentic sample analysis. This application note introduces a novel way users can customize data review within the scientific software for use in a routine environment.
Demonstrate accurate and facile review of HRMS data for determining the presence of targeted and unknown masses of interest in a spinach sample compared to a blank spinach sample.
Multi-analyte screening methodologies are essential for monitoring food and environmental samples across the globe. The goal of these methods is to eliminate the compliant samples and to identify the non-compliant samples for subsequent confirmation and quantification. Sensitivity must be in line with the relevant regulatory limits for residues in complex matrices. Also, a method must be validated in accordance with legislative requirements. This method would ideally be rapid, cost effective and a streamlined process, from sample preparation to reporting results.
To date, LC-MS/MS or GC-MS/MS tandem quadrupole technologies meet the requirements above and currently exist as the de-facto technique used to perform these analyses. However, with a constantly increasing number of analytes being added to monitoring and watch lists, the scope of a typical screening method is being extended. In addition, requests to screen for compounds beyond a target list are becoming increasingly common. As a result, many laboratories are progressing towards high-resolution mass spectrometry (HRMS) screening techniques that, in theory, can monitor for an unlimited number of targets at the same time as providing information to help discover unknown compounds or metabolites of interest.
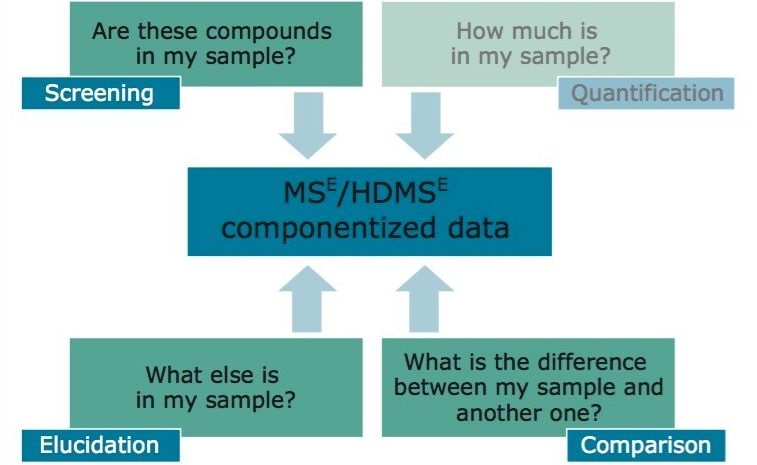
The ease of use and efficacy of a non-targeted, data independent, analysis type (MSE and HDMSE),1 coupled with a state-of-the-art scientific information system (UNIFI) for multi-analyte screening in food and environmental samples is demonstrated with this case study involving an authentic sample analysis. This application note will focus on introducing a novel way users can customize data review within the scientific information system in a routine environment. Details will include how to establish a concise, rapid, facile, and consistent approach to reviewing HRMS data to potentially answer the four questions listed in Figure 1 with a single processing step.
Vials containing spinach extract and spiked spinach extract at 1.0 g/mL matrix in 100% acetonitrile (ACN), prepared using QuEChERS, was supplied by a collaborator. The sample was diluted 1:1 with water, resulting in a concentration of 0.5 g/mL matrix. A 5-µL injection was performed. A non-targeted, data independent analysis, (MSE)1 was collected and processed in UNIFI. A previous application note2 details the parameters used to collect the non-targeted data set and highlights the importance of data compoentization3 to enable facile, consistent, and rapid interrogation of the complex data set produced.
The componentized data was interrogated against a target list of 529 agricultural residues and for unknown masses of interest. Non-targeted masses of interest were elucidated using a batch elucidation tool. This analysis focuses on the qualitative accurate mass screening, binary comparison, and unknown screening capabilities of Waters® Pesticide Screening Application Solution, answering three (highlighted) of the four questions shown in Figure 1.
The collaborator was confident that the spiked spinach sample would contain pesticide residues not on the target list. For the non-targeted residues, the collaborator wanted to assess how these residues were discovered and elucidated. A detailed description of how this was done is the focus for the Workflow Step Spotlight at the end of this application note.
The aim of these case studies is to show how a user can get from injection of a sample to an accurate report in a quick, efficient, systematic, and reproducible way using workflows, views, and filters within UNIFI. The workflow used for this qualitative analysis is shown in Figure 2. A workflow (left) is a series of steps which allows users to review HRMS data concisely and consistently, with each step consisting of a customizable filter and view.
The information displayed allows the user to make rapid decisions to questions listed in Figure 1.
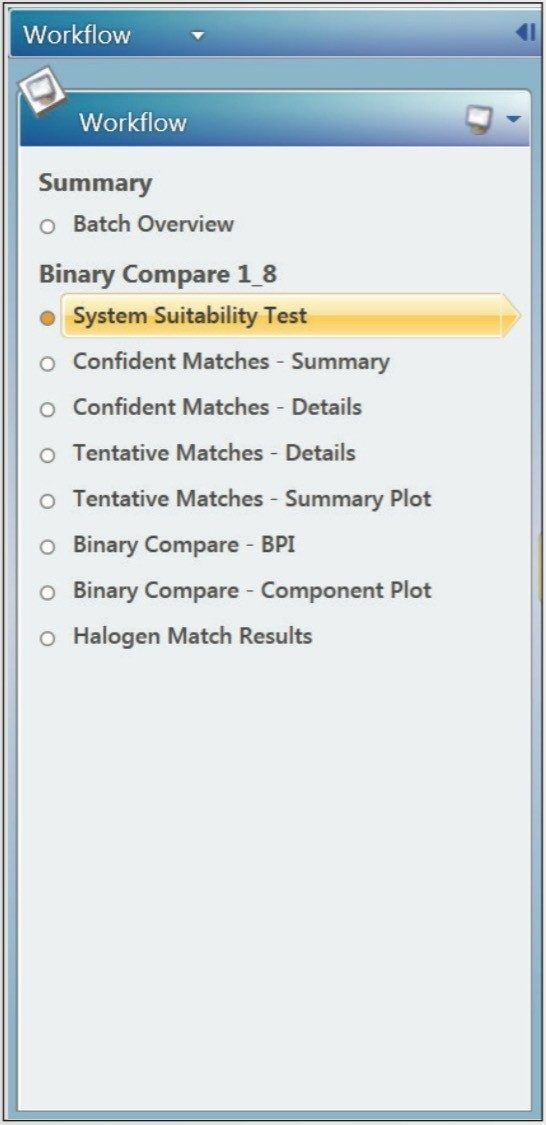
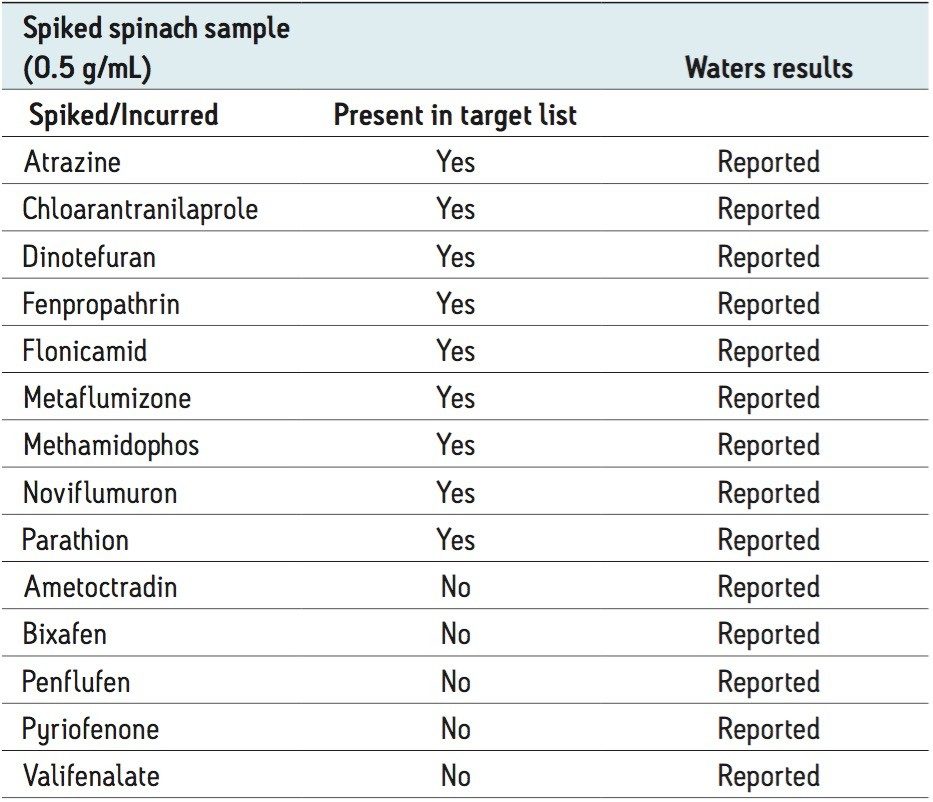
Table 1 shows the list of compounds spiked into the spinach sample as provided by the collaborator. Following data review, all compounds spiked into the spinach sample by the collaborator were reported. Nine compounds were present from the target list of 529. Five compounds of interest, not present in the target list, were discovered using a binary comparison with the blank spinach sample or by using the halogen match tool within UNIFI. Elucidation of these five masses of interest was performed using the discovery tool in UNIFI, which is essentially a batch elucidation tool.
Sensitivity – The use of MSE provides unbiased, non-targeted data sets with sufficient sensitivity to detect precursors and product ions for pesticides at concentrations below their MRL.
Speed – Scan rates for collecting a comprehensive MSE data are set according to the peak width of a developed UPLC method. The fast duty cycle allows a user to capture sufficient points across the chromatographic peak for both precursor and product ion channels in a single injection in order to maximize identification criteria and quantification results.
Selectivity – Apex 3D peak selection and componentization increases specificity and enables a user to interrogate data for targeted, non-targeted, and unknown masses of interest in a complex sample, without additional processing of raw data.
Efficacy – The use of filters, workflows, and views presents a consistent, concise, and comprehensive review of large data sets in a routine environment to enable a user to get from injection to accurate results fast.
Workflow Steps 6–8: Non-Targeted (Unknown) Screening Using Binary Compare and Halogen Match Features
Componentization of the data ensures that all candidate masses of interest are included in the same data set for interrogation, either through a list of targets, or for unknown masses of interest. There is no additional processing required to search for unknowns. All features (candidate masses) are extracted via componentization.
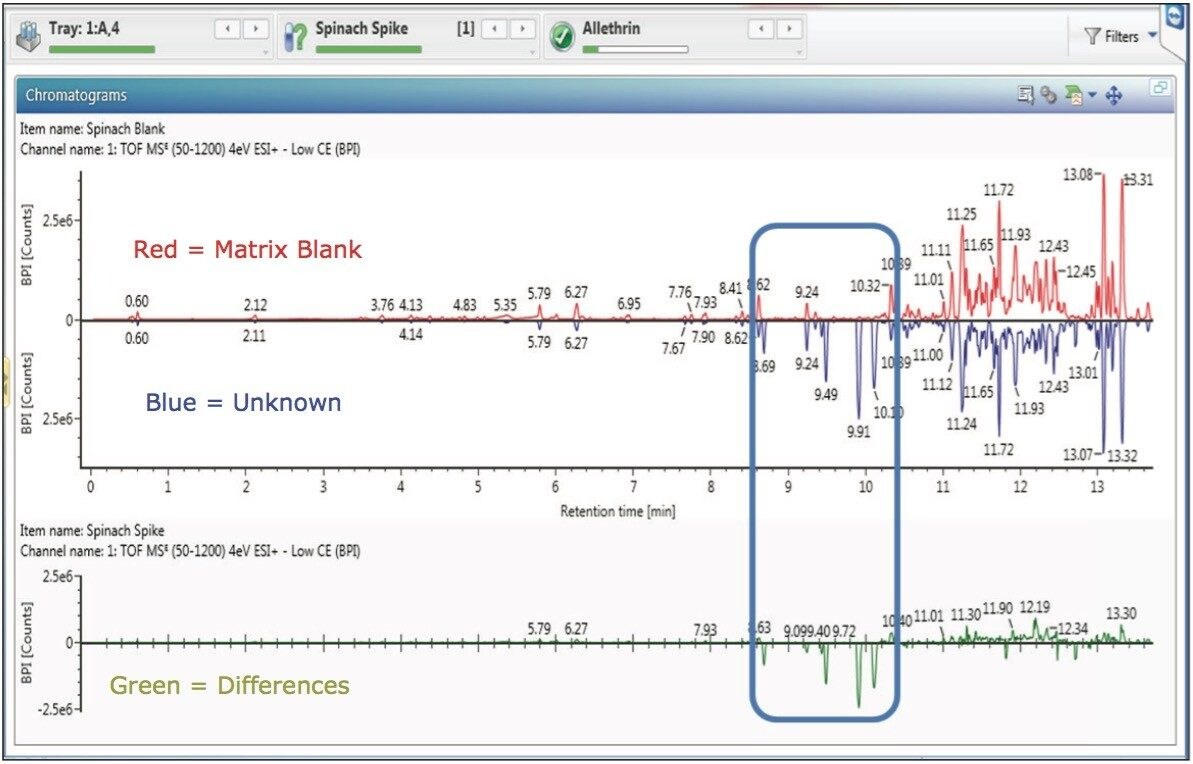
There are several ways to compare a reference sample versus an unknown sample in UNIFI. Large differences are easily displayed using base peak intensity (BPI) binary compare functionality (Figure 3).
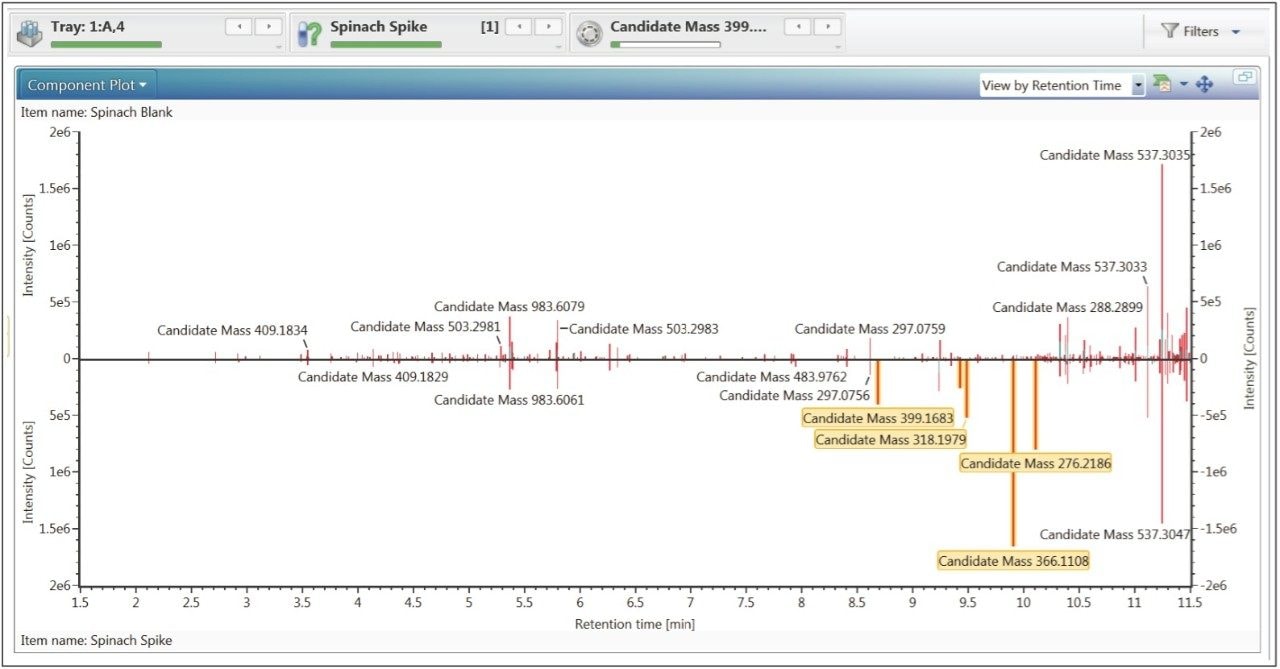
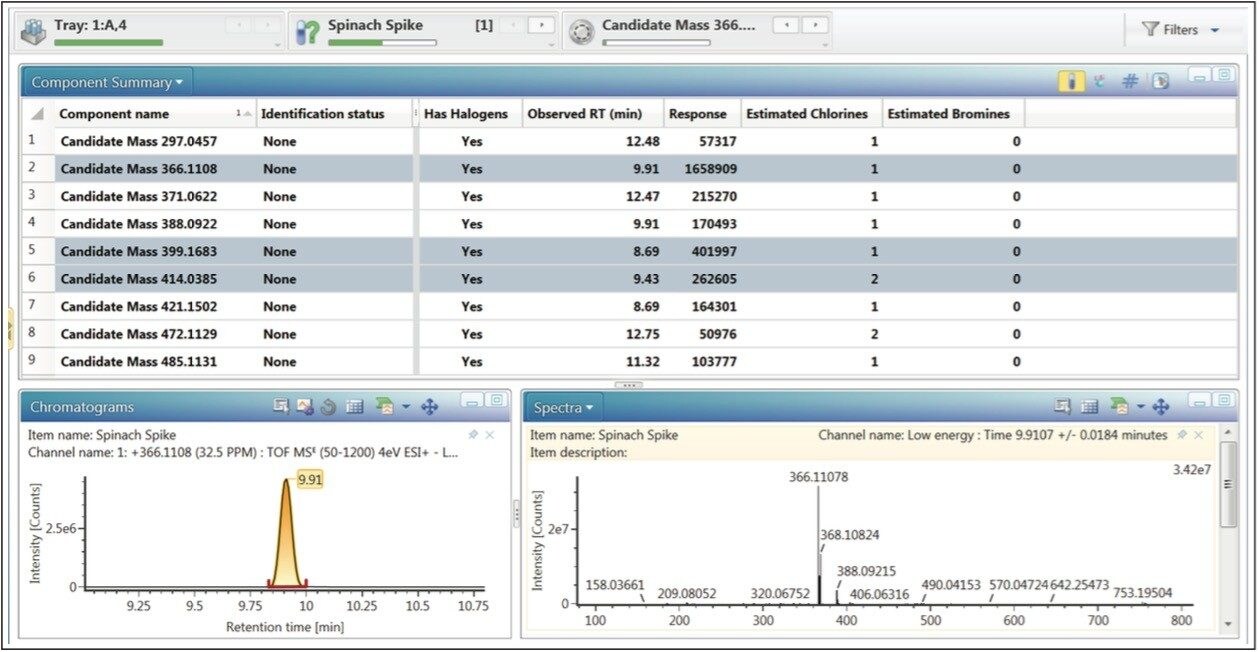
Unknown masses of interest can also be discovered using the halogen match function (Figure 6). During the single processing stage, every non-identified mass is assessed for the presence of chlorine and bromine atoms using mass difference and isotopic intensity. This workflow step uses a simple filter to highlight potential halogen containing masses above an intensity threshold, within a defined mass range. The information chosen for display is a component summary, an extracted ion chromatogram and spectral information (Figure 6). The component summary highlights information such as retention time, response and proposed number of chlorine and bromine ions. The spectrum window allows the user to quickly evaluate each potential halogen containing compound. The chromatogram window shows an extracted ion chromatogram (XIC) of a selected candidate mass. There are three masses highlighted in Figure 6. This means that three of the five masses highlighted in the binary comparison (Figure 5) also satisfy the criteria for the halogen match tool. Other tools (not shown) within UNIFI which enable discovery of unknown masses of interest are common fragment, mass defect and neutral loss. Full multivariate analysis capabilities (not shown) are also available for complete unknown screening experiments.
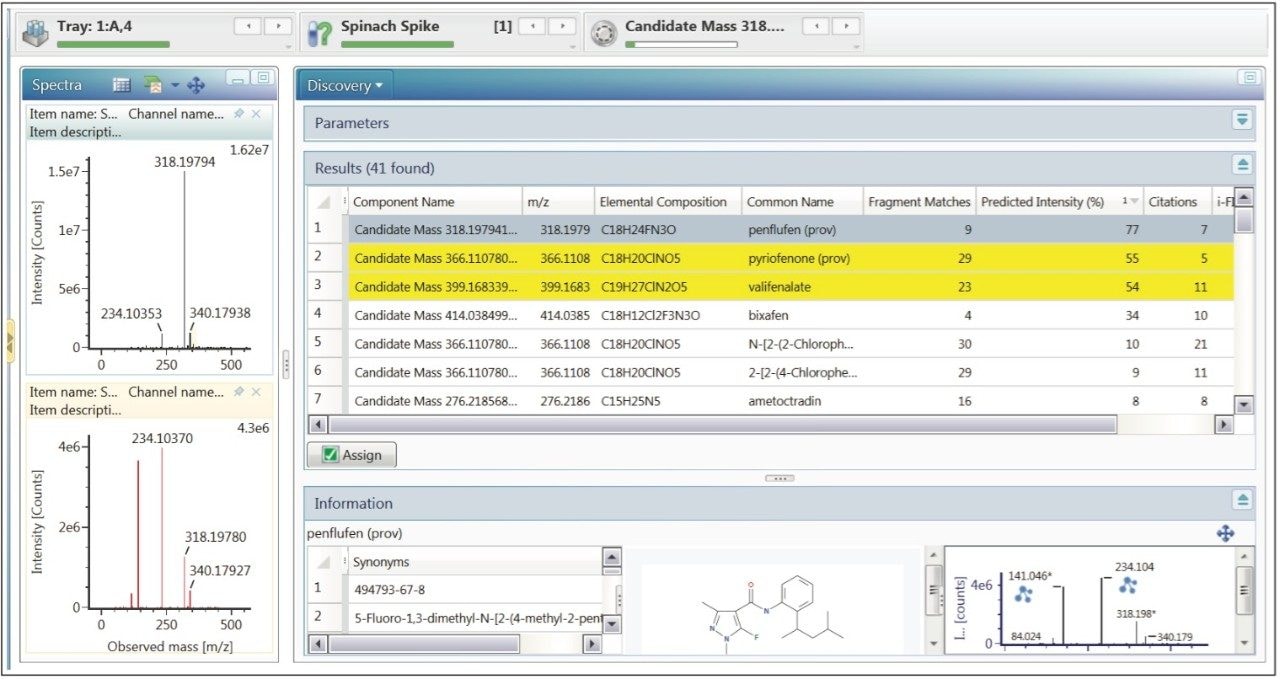
A wide range of elucidation tools are available within UNIFI to help identify unknown masses of interest. One of which is the Discovery Tool, which allows a user to submit multiple masses for elucidation as a batch. Primarily, Elemental Composition is performed on selected masses of interest. Any proposed formulae that meet the set criteria are then submitted to a library search. This could be an in-house library within UNIFI, an individual or selection of libraries within ChemSpider4 or the entire ChemSpider library. A mol file is downloaded for all potential library matches and fragment match is performed as the final part of the batch elucidation. The Discovery Tool settings are explained in more detail below.
Discovery Tool Settings:
Results from the submission of five masses of interest (highlighted in Figure 5) to the discovery tool are shown in Figure 7. The results table on the right in Figure 7 shows all of the predicted elemental compositions with the database search results and fragment match information for the five masses of interest. These results can be sorted by column header and in this case they have been ranked using predicted intensity. The top hit is highlighted for candidate mass 318.1979. This candidate has the predicted elemental composition of C18H24FN3O with a database search result of penflufen. The structure of penflufen from the database was used to perform a fragment match in which nine ions from the product spectrum were matched within a mass error of 2 mDa. This yielded a predicted intensity match of 77% of the spectral peaks present in the high energy data, which provides good confidence in this result. Spectral information for the precursor and product ions of the highlighted candidate mass is shown on the left hand side of Figure 7.
Following review of the discovery tool results a user can select the assign button and include these potential identifications in the final report.
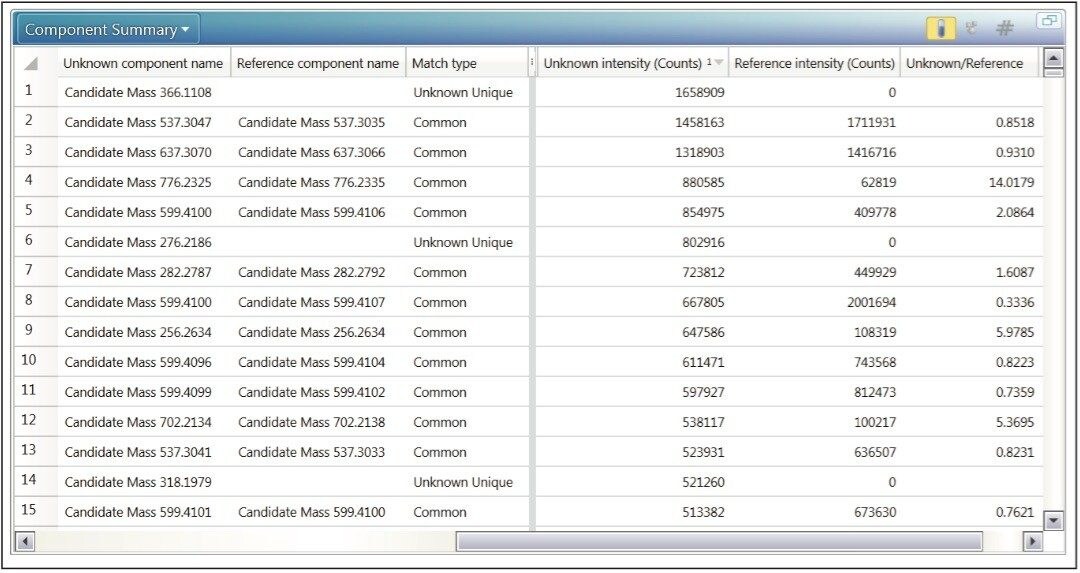
A user can also choose to display information in tabular form. ‘Unknown Unique’, ‘Reference Unique’ and ‘Common’ masses of all intensities are displayed when using the Component Summary to review data during the comparison of two samples (Figure 4).
With the application of a simple filter a user can focus on ions of interest. For example, only display masses that are unique to the unknown sample, that are between 100–500 Da and have a response greater than a defined value.
The binary compare Component Plot (Figure 5) displays component masses as sticks. This allows a user to instantly observe and select masses of interest that are different in the unknown sample compared to the reference. Once selected, these masses of interest (highlighted in yellow boxes) can then be sent directly to the elucidation toolset.
Essentially, with an MSE acquisition, data componentization, and UNIFI, a user can investigate, sort and display data for rapid reporting.
The use of halogen match, binary compare, and batch elucidation enabled identification of the five unknown compounds spiked in by the collaborator. These compounds (ametoctradin, bixafen, penfluen, pyriofenone, and valifenalate) were not disclosed by the collaborator prior to the analysis but were found to be correct upon final review of the analysis.
720005608, February 2016