
This application note presents an electrospray (ESI), positive mode, natural products library. The increased use of LC-MS-based methods to profile medicinal plants/herbal remedies and complex samples has become more pivotal in phytochemical profiling; but remains a challenge due to sample complexity.
The use of the ion mobility mass spectrometry (MS) collision cross sections (CCS) as an additional cumulative metric has been discussed previously.1-5 CCS measurements have been shown to be robust and reproducible.6 The use of CCS for small molecule analysis has increased across multiple research areas including pharmaceutical (e.g., metabolism, metabolomics, lipidomics) and food safety (e.g., veterinary drugs, mycotoxins, steroids, steviol glycosides, natural product screening, natural toxins). In these research areas, generation of CCS searchable libraries allows the application of a CCS metric which can increase the specificity of identification, and consequently can be used to decrease the false detection rate.
A strategy has been developed to facilitate the efficient and robust generation of mass spectrometry libraries that incorporate precursor/product ions, adduct ions, retention times, and CCS values. Briefly, the library building strategy utilizes analytical standards, which are screened in triplicate, using both positive and negative ion electrospray ionization. Average CCS values (including those of adducts) and product ions are extracted from the processed data to produce analyte-specific, MS library information.7
This application note presents an electrospray (ESI), positive mode, natural products library. The increased use of LC-MS-based methods to profile medicinal plants/herbal remedies and complex samples has become more pivotal in phytochemical profiling; but remains a challenge due to sample complexity.4
Green tea extract (p/n: 186006962)
33 mg was dissolved in 2 mL of methanol/water 1/3 solution, and diluted further to a working concentration of 8.25 mg/mL for analysis.
|
LC conditions |
||
|---|---|---|
|
System: |
ACQUITY UPLC I-Class |
|
|
Detection: |
Ion mobility mass spectrometry |
|
|
Vials: |
LCMS Certified Clear Glass 12 × 32 mm Screw Neck Total Recovery Vial, with Cap and Pre-slit PTFE/Silicone Septa, 1 mL volume, (p/n:600000671CV) |
|
|
Column: |
ACQUITY UPLC BEH C18, 1.7 μm, 2.1 × 100 mm (p/n: 186003950) |
|
|
Column temp.: |
45 °C |
|
|
Sample temp.: |
10 °C |
|
|
Injection volume: |
10 μL |
|
|
Flow rate: |
0.75 mL/min |
|
|
Mobile phase A: |
0.1% formic acid in water |
|
|
Mobile phase B: |
0.1% formic acid in acetonitrile |
|
|
Gradient: |
0–1 min isocratic (98:2.0 [A:B]), 5 min (95;5), 10 min (80:20), 13 min (70:30), 15 min (20:80), 15.1 min (98:2.0), and 17 min (98:2.0) |
|
System: |
SYNAPT G2-Si |
|
|
Ionization mode: |
ESI+ |
|
|
Capillary voltage: |
3 kV |
|
|
Cone voltage: |
30 V |
|
|
Desolvation temp.: |
550 °C |
|
|
Source temp.: |
150 °C |
|
|
Acquisition range: |
m/z 50–1200 |
|
|
Acquisition rate: |
10 spectra/s |
|
|
Lockmass: |
Leucine enkephalin (C28H37N5O7 (m/z 556.2766 +ve)) |
|
|
Collision energy: |
HDMSE low collision energy 4 eV and high collision energy ramp (30 to 75 eV) |
|
|
MS resolution: |
20,000 resolution full width half maximum (FWHM) at m/z 556 |
|
|
IM resolution: |
≈40 Ω/ΔΩ (FWHM) |
|
|
IMS parameters: |
Default IMS screening |
|
|
parameters include: |
T-Wave Velocity Ramp = Start: 1000 m/s End: 300 m/s, T-Wave Pulse Height = 40 V and a gas flow of helium 180 mL and nitrogen 90 mL (buffer gas) for the respective gas cells was used, giving an IM cell pressure of ~33.2 mBar |
|
|
Calibration: |
IMS/ToF Calibration Kit (p/n: 186008113) |
|
|
Chromatography software: |
MassLynx v4.1 SCN 916/924 |
|
MS software: |
MassLynx v4.1 SCN 916/924 |
|
Informatics: |
MassLynx data post processed using UNIFI v1.92 |
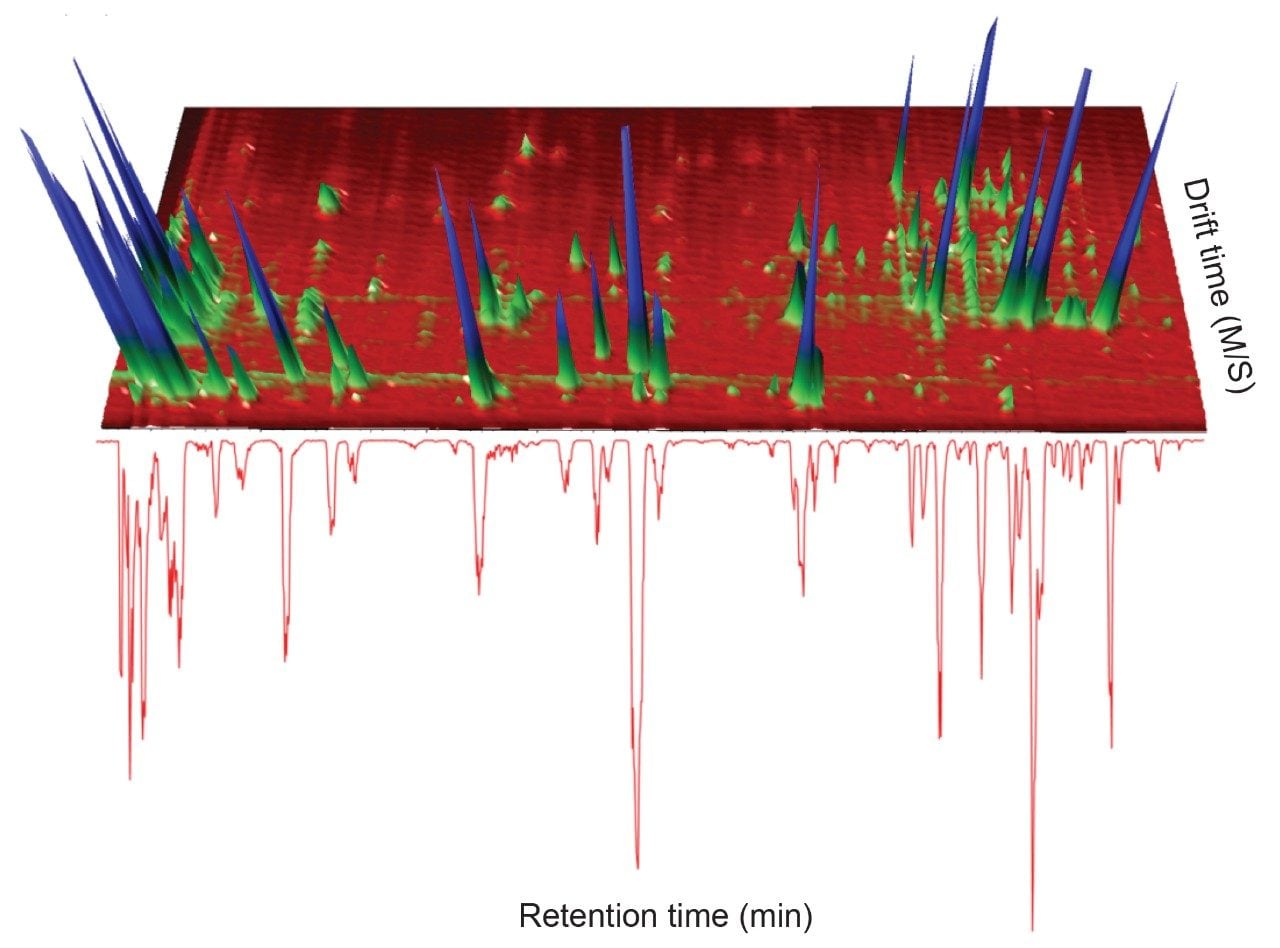
A positive ion mode, mass spectrometry library incorporating precursor/product ion, retention time using a standard gradient, and CCS values has been developed. A green tea extract was screened to assess the utility of the library. The UPLC-MS BPI chromatogram obtained, and the corresponding orthogonal ion mobility separation, is shown in Figure 1, where it is evident that coeluting analytes can be deconvoluted using ion mobility (separation due to shape, charge, and mass).8,9
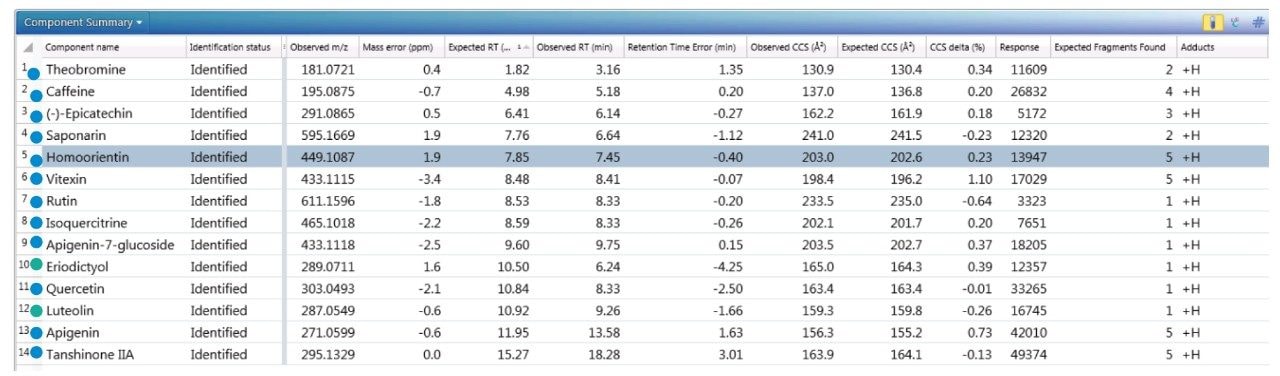
The component summary for analytes identified is shown in Figure 2. Literature searches have confirmed that many, if not all, of the identified components are known to be present in green tea. However, it should be noted, confirmed detection/identification is based on generation of a match to a constituent of the library generated. 7-C glycosylated flavones (13), flavanols (12), flavan-3-ols (2,4), purine alkaloids (1,3), and flavonoids (5–11) have been identified. 6-C/8-C glycosides (homoorientin/orientin and isovitexin/vitexin) were previously determined to be present in black tea and vitexin in green tea.10,11 From the manual interrogation of the exact mass extracted mass chromatogram of orientin, evidence of a shoulder due to another isomeric species was observed.
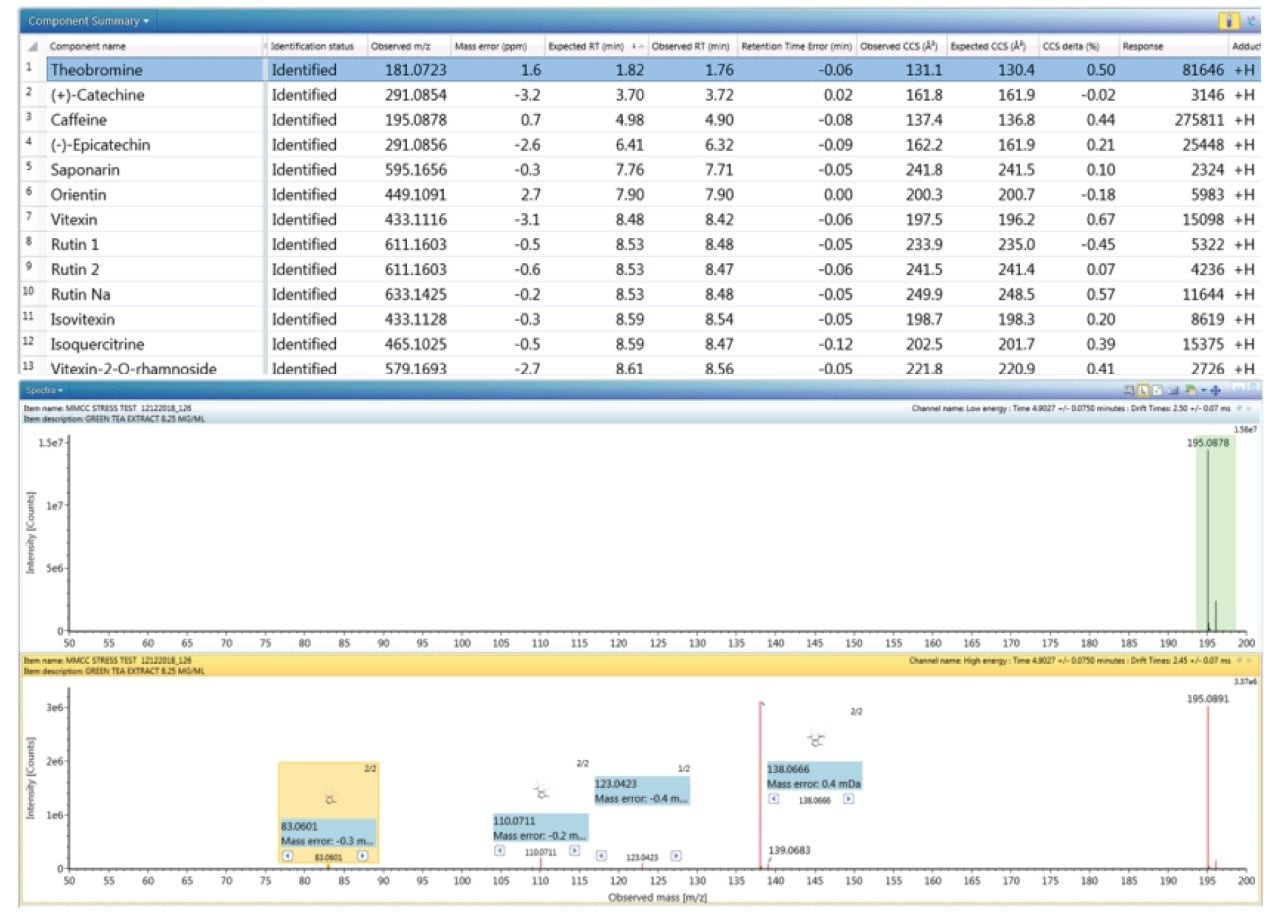
Figure 2 also shows a set of identifications made with mass accuracy <3.2 ppm, and <1% variation for CCS. The CCS metric provides an additional identification point for analytes of low abundance where there is insufficient ion intensity for product ion formation. The product ions generated are highly specific, because they are both retention time and drift time aligned, which is an enhancement compared with mass spectrometry techniques that produce retention time aligned fragment ions alone. The ion-mobility product ion spectrum of caffeine detected in green tea is presented in Figure 2. Excellent mass accuracy of the product ions is also llustrated (within 0.5 mDa), when compared to the expected product ions of caffeine in the natural products library. Caffeine was here identified using seven identification points, precursor ions, four product ions, retention time, and its CCS (0.4% delta).
To further illustrate the utility, robustness, and confidence that can be obtained when using a natural products, multi metric mass spectrometry library, green tea extract was fortified with a series of known analytes. These analytes are shown in Figure 3, where the component summary for identified unknown analytes and known analytes are also presented. All fortified analytes were detected and identified correctly. Fortification of the 6-C/8-C glycosides (homoorientin/orientin and isovitexin/vitexin) confirmed their presence within the green tea extract. Identification of such isomers can prove to be a challenge. In Figure 3, it can be seen that homoorientin is identified at 7.85 minutes and orientin at 7.90 minutes, with only 0.05 minute difference in observed retention time; however, the incorporation of CCS into the library has enabled the observed difference in CCS to be utilized, and the relative values are sufficient to ensure that the correct identification assignment can be made. This is relevant whether the library is used within UNIFI or Progenesis QI Software.
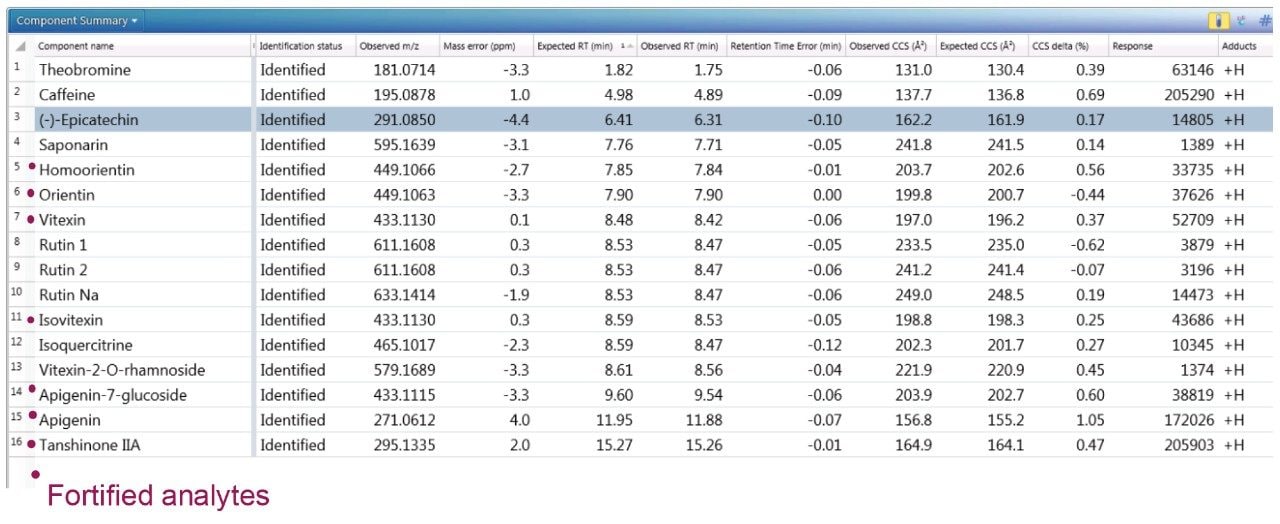
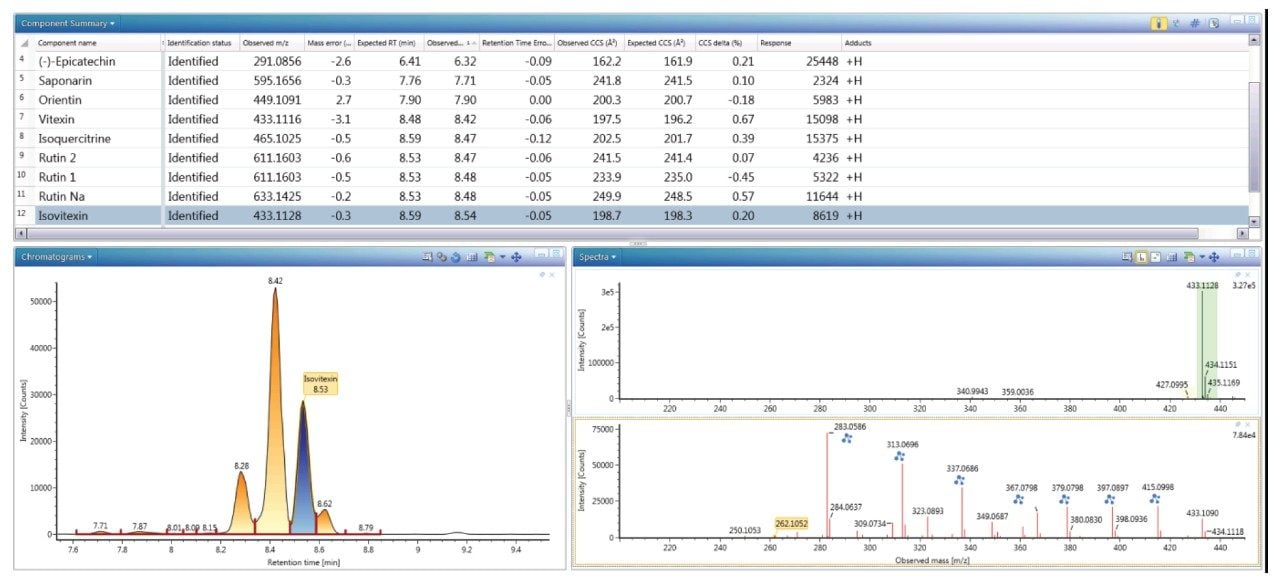
Figure 4 shows the extracted, exact mass chromatogram for m/z 433.1128, where a series of ten minor/major peaks were observed with four isomers within a retention time window of 0.4 minutes. When performing natural products analysis, the analyte concentration in the extracts can vary significantly. Frequently many isomeric species are present, which cannot be identified specifically using only accurate mass measurement alone. Subtle variations in retention time and analyte concentration will impact the ability to identify such isomeric species with confidence. The challenges and successful characterization/ identification of isomeric species has previously been described using cumulative specificity of ion-mobility CCS, retention time, and ion-mobility product ions.4
The advantage of enhancing the specificity of the mass spectrometry library using CCS is illustrated in Figure 5, where green tea extract has been analyzed using an alternative chromatographic method to that used to produce the natural products library (Waters Application Note, p/n: 72004837EN). A retention time window of 10 minutes was utilised rather than 0.2 minutes, and the largest candidate response within the retention processing window is reported in UNIFI’s component summary. The strategy employed in data processing utilizing only CCS and m/z as the screening parameters can be used as a highly time efficient tool to investigate/compare analysis results where different chromatographic methods are employed. As can be seen from Figure 5, impressively, most analytes identified in the component summary in Figure 3 have been identified, also two additional analytes were identified, which were not previously documented in the literature as being present in green tea. This approach illustrates that more flexibility can be attained from the utility of mass spectrometry libraries by increasing the cumulative specificity and selectivity of the identification points within the library.12 Additional identification points/specificity can also result from multiple adducts, and the formation of protomer’s (charged isomers) or conformers,3 where more than one ion mobility peak is observed for a single analyte. This can be illustrated for rutin, for which 3 CCS values are obtained, comprised of 235 Å2 (conformer 1), 241.4 Å2 (conformer 2), and 248.6 Å2 ([M+Na]+), producing a highly specific CCS fingerprint, which has been uniquely incorporated into the natural-products, mass spectrometry library.
In the natural products library, luteolin 3,7 luteolin-diglucoside (C27H30O16) has an expected retention time of 8.39 minutes, compared to 8.54 minutes for rutin (C27H30O16), both within the 0.2 minute retention time tolerance window set for data processing. However, the unique rutin CCS fingerprint has facilitated its differentiation from luteolin 3,7 diglucoside.
720006650, September 2019