
For research use only. Not for use in diagnostic procedures.
This application note reports an APGC-TOF MSE profiling approach and its application to metabolic fingerprinting of Arabidopsis.
APGC-TOF MSE with Progenesis QI is a valuable solution for metabolomics applications. The use of orthogonal information for the metabolite identification, including accurate mass, retention time, and theoretical or measured fragmentation, increased the confidence of metabolite identification while decreasing the number of false positive identifications.
Atmospheric pressure gas chromatography mass spectrometry (APGC-MS) provides molecular ion information, which is typically absent when traditional vacuum source (i.e., electron ionization) gas chromatography mass spectrometry (GC-MS) is employed. This application note highlights the use of APGC-MSE for analysis in metabolomics.
Gas chromatography coupled with mass spectrometry is a well established analytical approach to metabolomics. The most widely used ionization technique is electron ionization (EI), which produces highly fragmented spectra that are library searchable. The molecular ion in an EI spectrum is often absent or of very low abundance. The lack of molecular ion information, especially for derivatized compounds in complex mixtures, can lead to incorrect identification when using spectral matching alone.
Atmospheric pressure GC (APGC) is a soft chemical ionization technique that generates a mass spectrum in which there is minimal fragmentation and conservation of the molecular ion (Figure 1). Additionally, when APGC is combined with a time of flight mass spectrometer (TOF-MS) for exact mass MSE analysis, comprehensive precursor and fragment ion spectral data is obtained for every detectable component in a complex sample. GC-MSE offers a simple route to providing high selectivity and confidence for simultaneous identification and quantitation in a single analysis.
Here we report an APGC-TOF MSE profiling approach and its application to metabolic fingerprinting of Arabidopsis.
![Figure 1. A) An APGC System can be coupled to various Waters MS instruments, including the Xevo TQ-S and the SYNAPT G2-S. The changeover from UPLC to APGC takes less than five minutes. B) The APGC source consists of an ion chamber with a corona pin inside. The GC column enters the source via a heated transfer line. Corona discharge creates nitrogen plasma within this region. Radical cations generated in this plasma interact and ionize the analyte molecules. The ions created are then transferred to the mass analyzer. C) Under dry source conditions the predominant method of ionization is charge transfer, generating molecular radical cations [M+•]. This method favours relatively non-polar molecules. D) When a protic solvent, such as water or methanol, is added to the source, the predominant ionisation is proton transfer, generating [M+H]+ ions. This method favors relatively polar molecules.](/content/dam/waters/en/app-notes/2015/720005298/720005298en-f1.jpg.82.15-14-683-969C.resize/img.jpg)
|
GC system: |
7890A GC |
|
Column: |
HP-5MS column, 30 m length, 0.25 mm I.D., and 0.25 μm film thickness (Agilent Technologies) |
|
Carrier gas: |
Helium 1 mL/min |
|
Temp. gradient: |
Initial 70 °C, 5 °C/min to 310 °C, hold 1 min |
|
Injection type: |
split mode (split 4:1) |
|
Injector temp.: |
230 °C |
|
Injection vol.: |
1 μL |
|
Make-up gas: |
Nitrogen 400 mL/min |
|
Transfer line temp.: |
310 °C |
|
MS system: |
SYNAPT G2-S HDMS |
|
Mode of operation: |
TOF-MSE |
|
Ionization: |
APGC |
|
Corona current: |
3 μA |
|
Cone voltage: |
20 V |
|
Source temp.: |
150 °C |
|
Cone gas: |
10 L/h |
|
Auxiliary gas flow: |
500 L/h |
|
MS gas: |
Nitrogen |
|
Acquisition range: |
50 to 1200 |
|
Transfer CE: |
Ramp 20 to 40 V |
Progenesis QI Software v1.0
MassLynx Software v4.1 SCN870
Arabidopsis thaliana seeds were grown under controlled conditions. Seedlings were harvested and polar metabolites were extracted and derivatized. The dried polar phase was methoximated for 90 minutes at 45 °C and trimethylsilylated for 30 minutes at 37 °C.
APGC-MS analysis of commercially available pure reference compounds of known plant metabolites was performed. Following the analysis, an in-house APGC reference database was created containing retention times, and accurate mass-to-charge ratio (m/z) for precursor and fragment ions (Figure 2). APGC provided abundant molecular ions with minimal fragmentation at low collision energy (Figure 2A). To add confidence to compound identification, collision energy was ramped from 20 to 40 eV in the high energy function to generate maximum information from fragment ions (Figure 2A). Due to the use of charge exchange chemical ionization, elevated collision energy data resulted in a spectrum similar to the traditional EI data (Figure 2B).

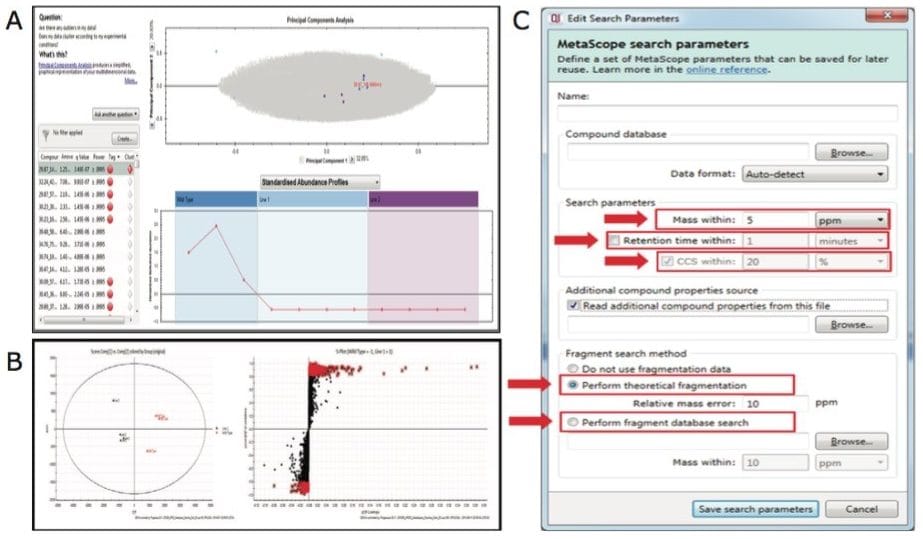
Plant extracts were analyzed using APGC-TOF MSE and raw data were imported to Progenesis QI Software for processing and analysis (Figure 3A and B). Multivariate statistical analysis highlighted the molecular differences between groups of samples (Figure 4A and B). The Progenesis QI search engine Metascope allowed us to query experimentally-derived or in-house databases. We were able to customize the search parameters for the metabolite identification according to multiple orthogonal measures such as mass accuracy, retention time and fragmentation matching (Figure 4C). Additionally, if ion mobility is activated, collision cross-section (CCS) data, which reflect the ionic shape of the metabolites in the gas phase, can also be mined for identification.
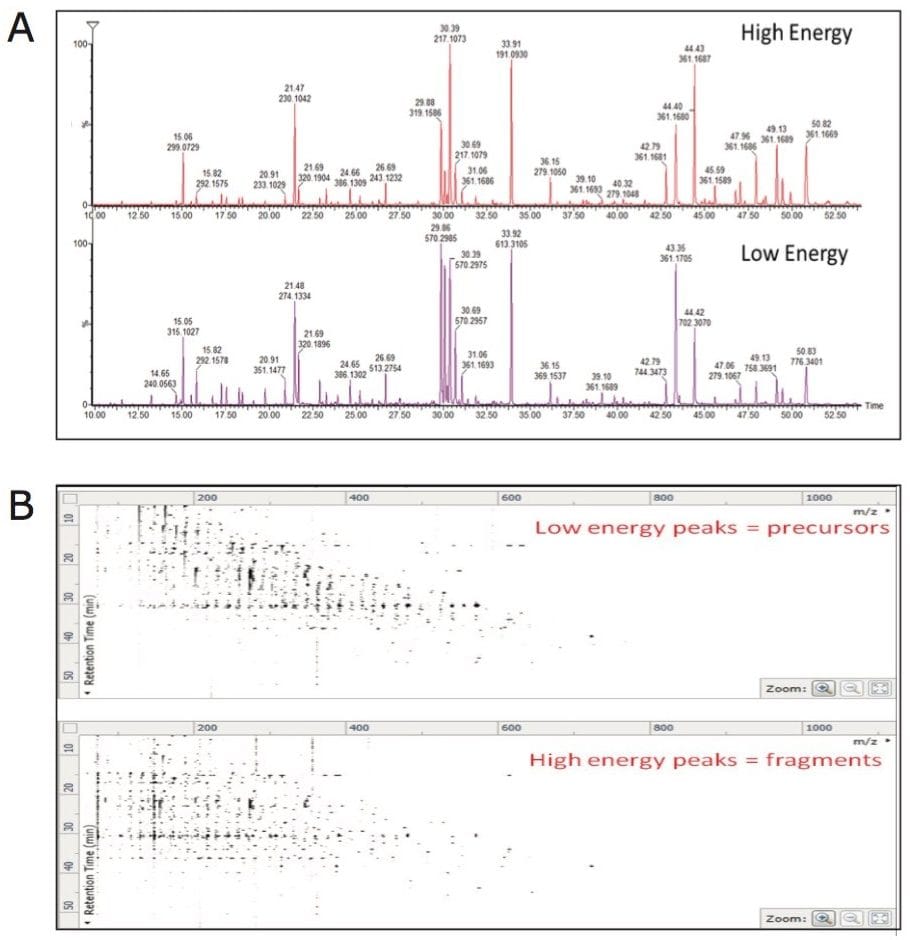
Sample data, generated with the low energy (precursor ion) spectrum and high energy (fragment ion) spectrum of each component, was searched against both the in-house and commercial mass spectrum libraries (Figure 5A and B). The identification score described the level of confidence obtained for each library hit based on accurate mass for precursor and fragment matching, retention time, and isotope ratios (Figure 5A and B).
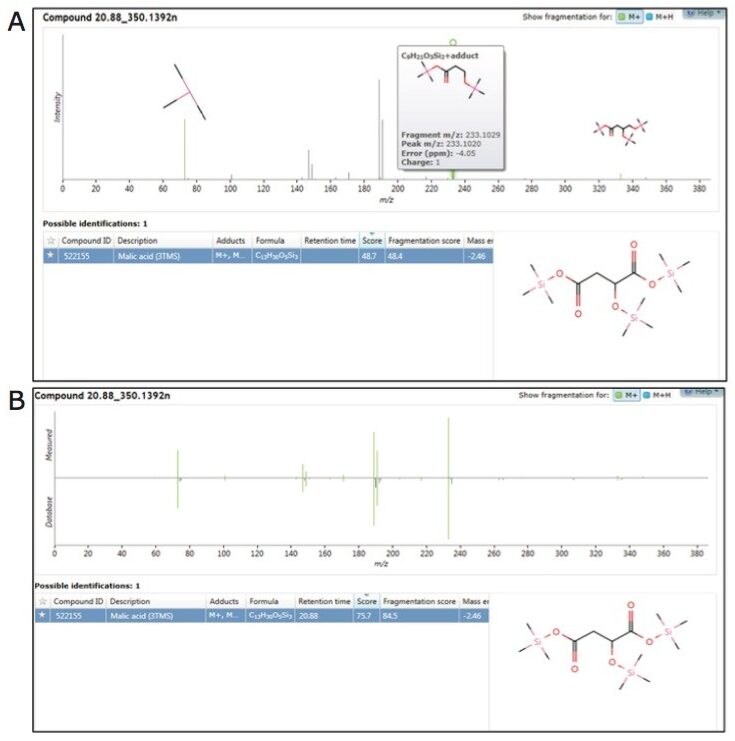
As structurally similar metabolites often co-elute, the concurrent acquisition of an intact molecular ion, along with fragmentation data for sub-structural determination, was particularly useful. In combination with accurate mass measurement, the molecular ion helps determine the limits of chemical composition, which can subsequently be used along with the fragmentation data for more detailed and specific structural elucidation of both known and unknown metabolites.
A summary workflow for the APGC-TOF-MSE approach:
APGC-TOF MSE with Progenesis QI is a valuable solution for metabolomics applications. The use of orthogonal information for the metabolite identification, including accurate mass, retention time, and theoretical or measured fragmentation, increased the confidence of metabolite identification while decreasing the number of false positive identifications.
720005298, February 2015