For research use only. Not for use in diagnostic procedures.
In this work, we demonstrate their use with MALDI imaging in studying the changes over time in the proteome on xenograph tissues sections after administration of an anti-cancer drug.
New features have been implemented into Waters High Definition Imaging (HDI) 1.2 Software to allow multivariate data analysis, such as principal component analysis (PCA) and partial least squares discriminant (PLS-DA). These capabilities, integrated to the workflow, reduce the dimensionality of data and ease the comparison of multiple datasets. In this work, we demonstrate their use with MALDI imaging in studying the changes over time in the proteome on xenograph tissues sections after administration of an anti-cancer drug.
In biomedical research, proteomics has become an indispensable tool for the discovery of candidate biomarkers and drugs. Moreover, mass spectrometry imaging (MSI) enables researchers to determine the spatial distribution of proteins and peptides directly from a tissue section, without radioisotope labeling or tagging.
However, a MALDI imaging experiment can readily generate a vast amount of data, depending on tissue size and acquisition mass range. Both of these factors relate to the number of ion detections, number of pixels recorded, and the possible addition of ion mobility separation to improve the specificity of the analysis. Moreover, in the case of comparing multiple samples, such as different states of a disease or drug time-course experiment, data review complexity is multiplied, amounting in tedious and time-consuming data review to identify molecular species changes.
|
Data acquistion |
|
|---|---|
|
System: |
MALDI SYNAPT G2 HDMS, positive mode |
|
Mass range: |
600-3,000 Da |
|
Laser: |
1 KHz solid state Nd:YAG laser (λ = 355 nm) |
|
Spatial resolution: |
150 μm (lateral) |
The obtained data were processed and visualised using HDI MALDI 1.2 Software. Regions of interest (ROIs) information were analysed using EZInfo (Umetrics Software). MassLynx Software was also utilized.
Four rats were injected subcutaneously with a cancer cell line to allow the growth of tumor mass, called a xenograph. The animals were administrated with the anti-cancer drug Dasanitib at a concentration of 30 mg/kg and scarified at different time points (control, 1, 3, and 6 hours). After tumour excision, frozen tissue sections from each animal were thaw-mounted onto a single glass slide. Tissue samples were washed with different baths of cold ethanol and chloroform. In situ trypsin digestion was performed by spraying a solution of trypsin directly onto the tissue and incubating overnight in a humidity chamber at 37 °C.
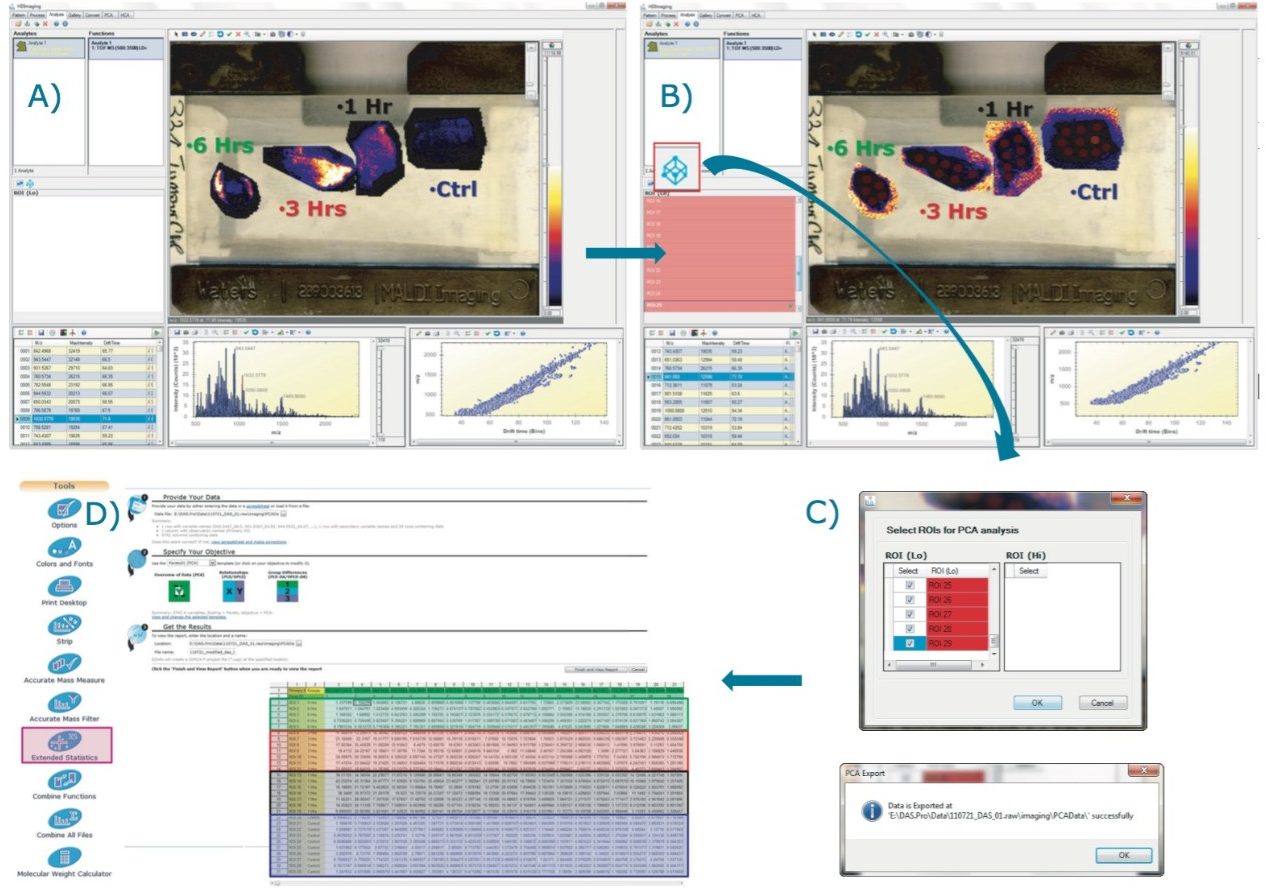
Post-digestion, several layers of matrix, α-cyano-4-hydroxicinnamic acid (CHCA) containing aniline in acetonitrile/water/TFA (1:1:0,1), were also sprayed directly onto the tissue samples. A single jpeg image of the four tissue sections mounted on the glass slide was taken using a flatbed scanner. Using High Definition Imaging (HDI) MALDI software, the visual image was co-registered, and areas to be imaged by MSI were defined. This allowed the overlay of the MS information and the visual image later in the analysis (Figure 2A).
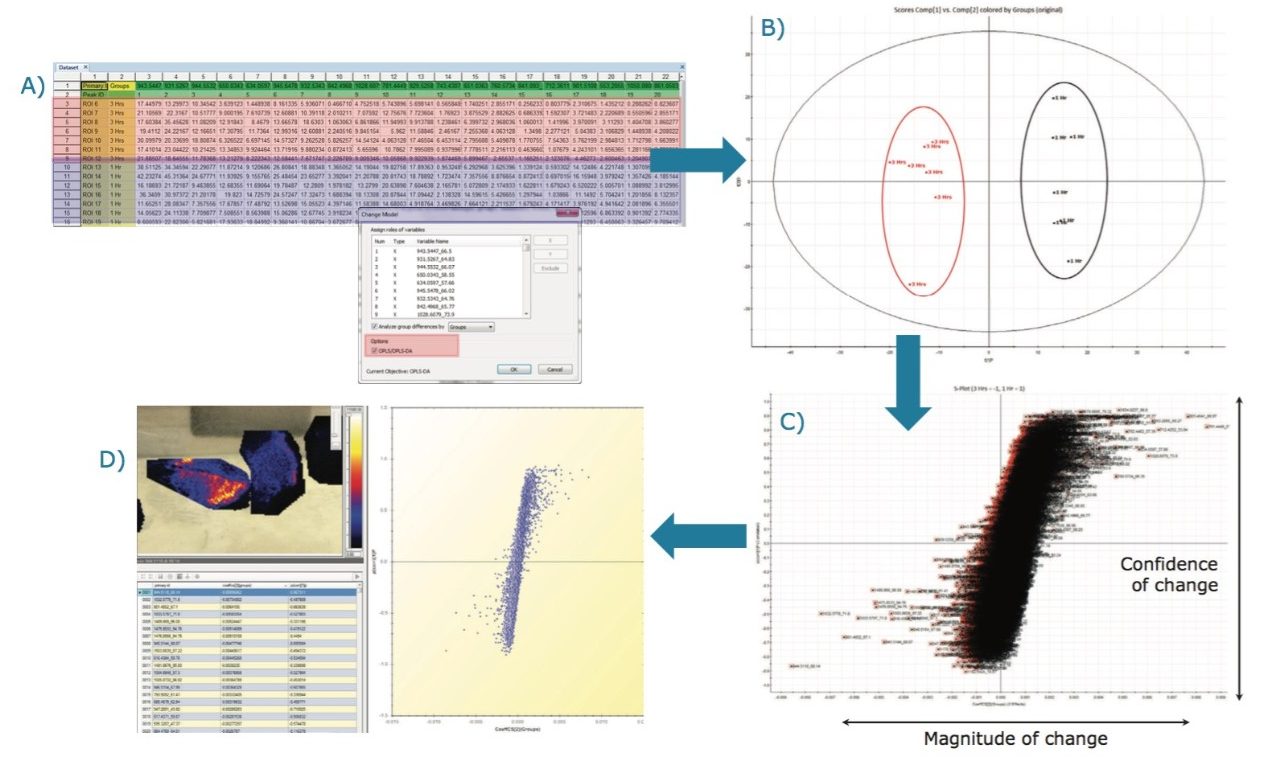
Figure 2A represents the processed data loaded in the Analysis tab of HDI Software. It is possible to define one or several regions of interest (ROIs) on the four tissue sections as shown in Figure 2B. The ROIs can be drawn using the free draw tool, eclipse (or round), and rectangle (square) options.
By clicking on the Umetrics icon as shown in Figure 2B, the intensities for each entry (m/z, dt) present in the processed data are averaged and TIC normalized across the pixels present within each defined ROI. The output is reported in a csv file (Figure 2C). EZInfo can be launched directly from MassLynx Software by clicking on the Extended Statistics icon (Figure 2D). The output txt file is loaded in EZinfo where the different groups can be specified, as seen in Figure 2D).
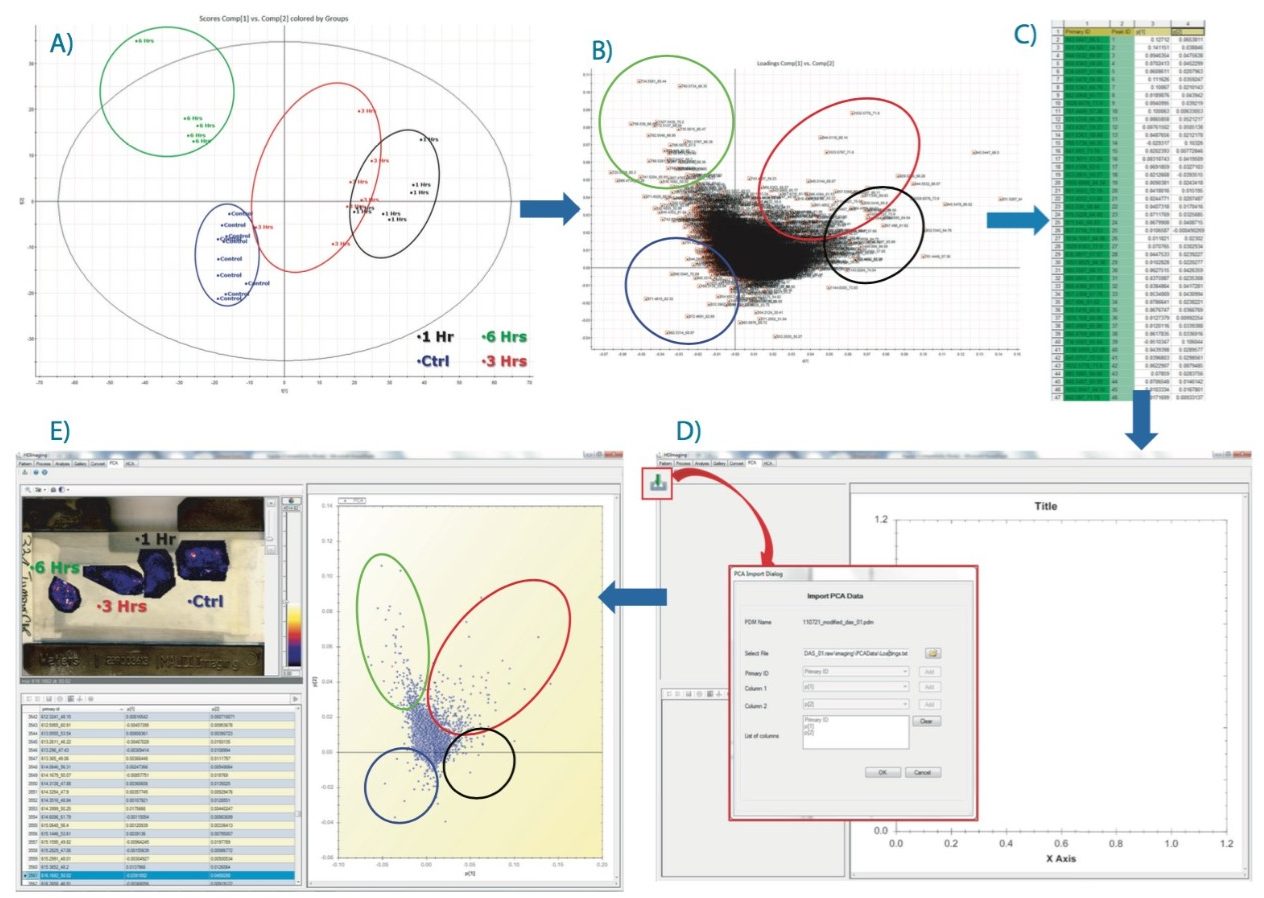
Following on from the data matrix loaded in EZInfo, as shown in Figure 2D, PCA is performed and the result scores plot displayed in Figure 3A. Each dot represents an ROI observation, which has been color coded to one of the four groups (i.e., one of the tissue section). Scores values for each group clustered well together.
A second data processing step was performed within EZInfo to obtain the loadings data for the analysis as demonstrated in Figure 3B. Each loading dot, representing a (m/z, dt) entry, can be selected and a list can be generated comprising peak IDs of each of the selected variables with its associated PCA coordinates (Figure 3C). This list can be imported back into the HDI Software following the route shown in Figure 3D. The loadings plot is recreated in HDI Software (Figure 3E) and the dataset queried to highlight ion images associated with variables from the loadings distribution.
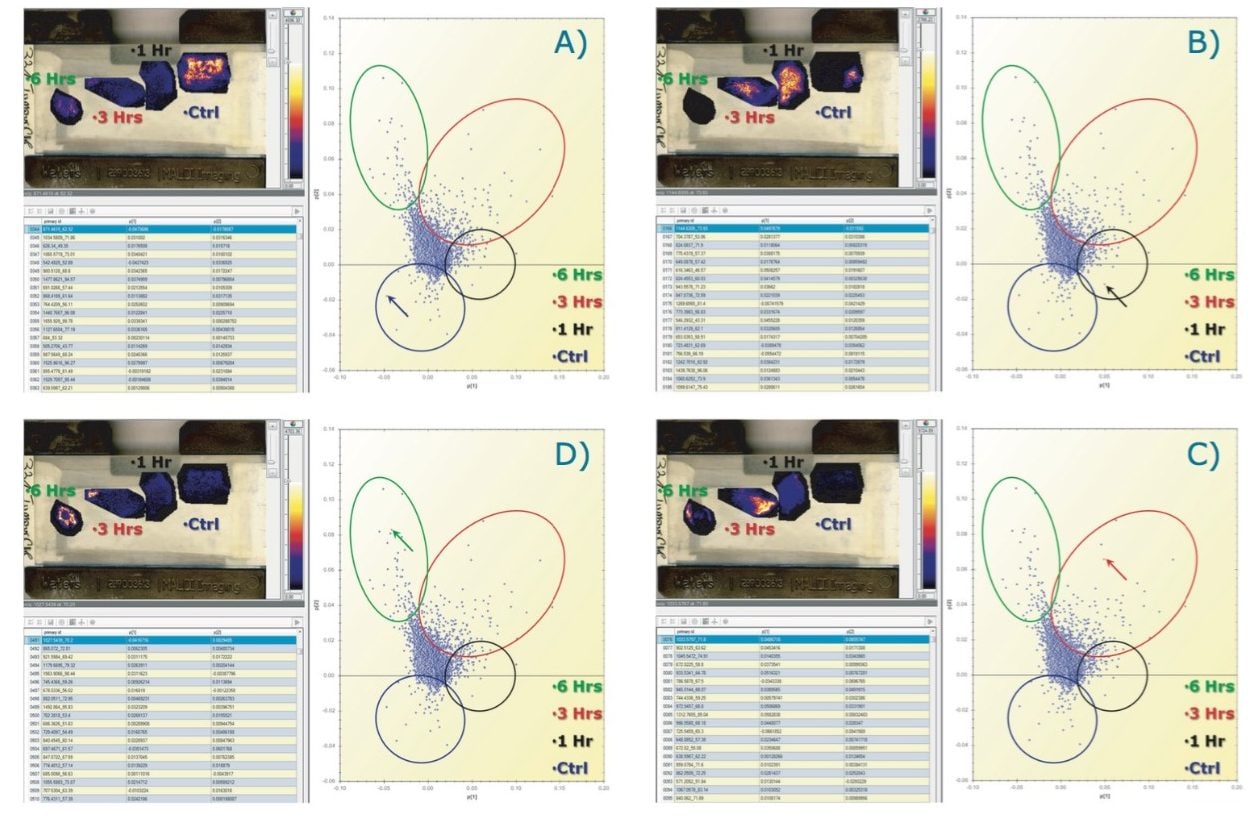
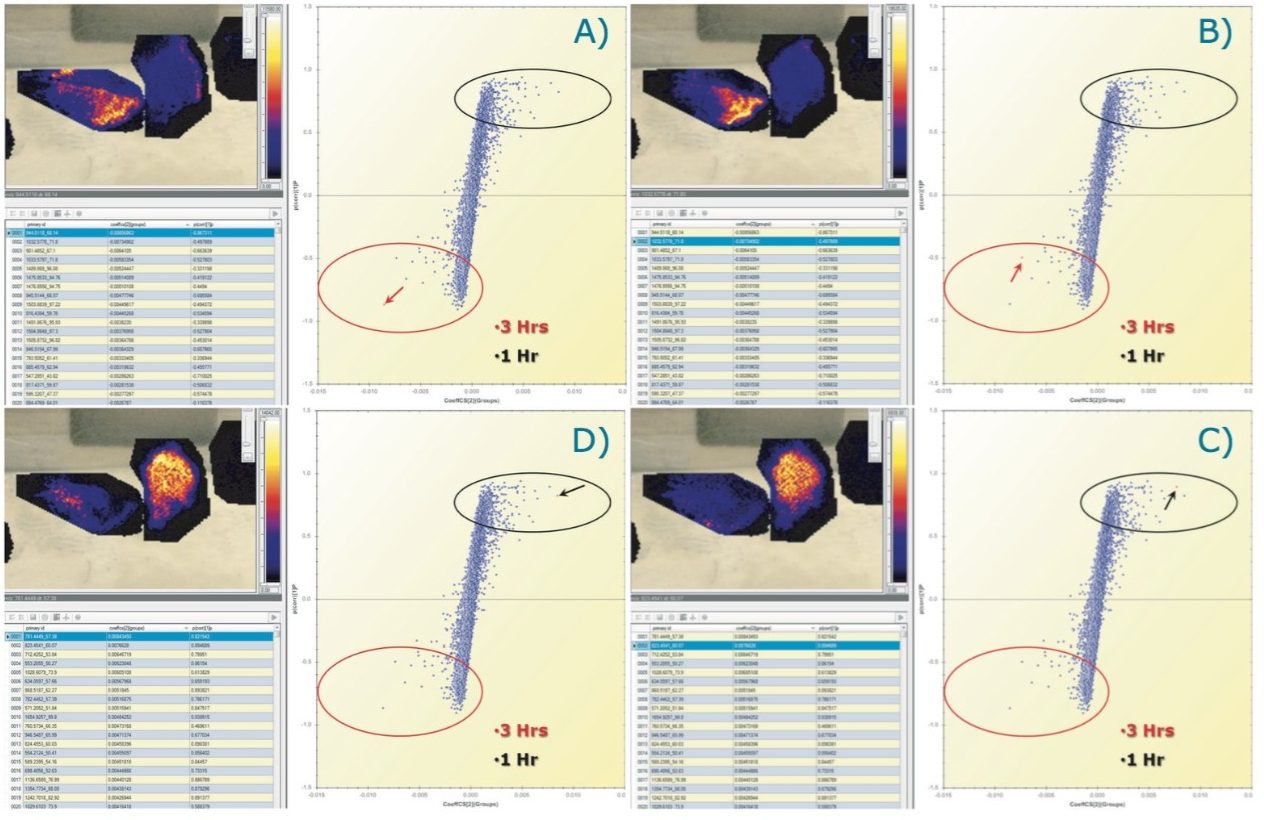
The latter is illustrated in Figure 4, where ion images of specific tryptic peptides are likely to distinguish the different conditions from the PCA experiments. Figure 4A shows the selection in the loadings distribution of variable (m/z_dt) 871.5_62, which is dominant in the control condition compared to the other tissue section images. Figure 4B shows the selection of (m/z_dt) 1144.6_74 that is more abundant in the 1 hour condition. Figure 4C shows the selection of (m/z_dt) 1033.6_72 that is more abundant in the 3 hours condition and Figure 4D shows the selection of (m/z_dt) 1027.54_70 that is more abundant in the 6 hours condition.
From the data matrix of (m/z_dt) analyte values, intensities, and group definitions shown in Figure 2D, it is possible to select only two groups, such as group 1 and 3 hours (Figure 5A), which were the two groups that were the least clearly differentiated in the unsupervised analysis.
An OPLS/OPLS-DA distribution was generated (Figure 5B) with its S-plot (Figure 5C) that illustrates confidence of change on the Y-axis and magnitude of change on the X-axis. As for the unsupervised analysis, the loadings data can be imported back into the HDI Software where analyte/ion images associations can be viewed (Figure 5D) and queried (Figure 6).
720004873, November 2013