This is an Application Brief and does not contain a detailed Experimental section.
This application brief illustrates that with careful method development, peptides can be measured at low limits of quantification (LOQ) in complex protein digest samples using properly configured MRM analysis.
***nanoACQUITY UPLC applications readily transfer to the ACQUITY UPLC M-Class System***
Easily achieve quantification of less than a femtomole from complex protein digest samples using a nanoACQUITY UPLC System coupled with a Xevo TQ MS.
The selectivity and sensitivity attainable using MRM data acquisition on a triple quadrupole mass spectrometer applied to the analysis of peptides in protein digests for the purpose of quantifying proteins in biological samples has attracted great interests in many areas of life science. These include verification of potential biomarkers in diseases such as cancer, and quantification of therapeutic proteins. In both of these cases, the likely matrix will be blood plasma or serum. This analysis will be successful if all of the following conditions are met:
Moreover, all sample preparation operations must be reproducible.
Five tryptic peptides for each of the four proteins comprising the MassPREP Protein Expression Mixture 1 Digestion Standard were selected for MRM analysis, and several MRM transitions were chosen for each peptide with the assistance of VERIFYE Software. Mixture 1 was analyzed with a Xevo TQ Mass Spectrometer with a nanoACQUITY UPLC System equipped with a 75 μm diameter column using 1 μL injections. The tryptic digest mixture was injected at 100 fmol/μL by itself and at concentrations ranging from 200 fmol/μL to 390 amole/μL in a matrix of 1 μg/μL of digested human serum proteins. The results were curated according to the criteria listed above.
Figure 1 shows unsmoothed chromatograms for the peptide SIGGEVFIDFTK derived from alcohol dehydrogenase (S. cerevisiae) in the simple mixture, and at three concentrations in the serum matrix. These chromatograms exhibit acceptable peak shape and are generally free of any interference, though some noise is visible between 50 and 52 minutes at the lowest concentration.
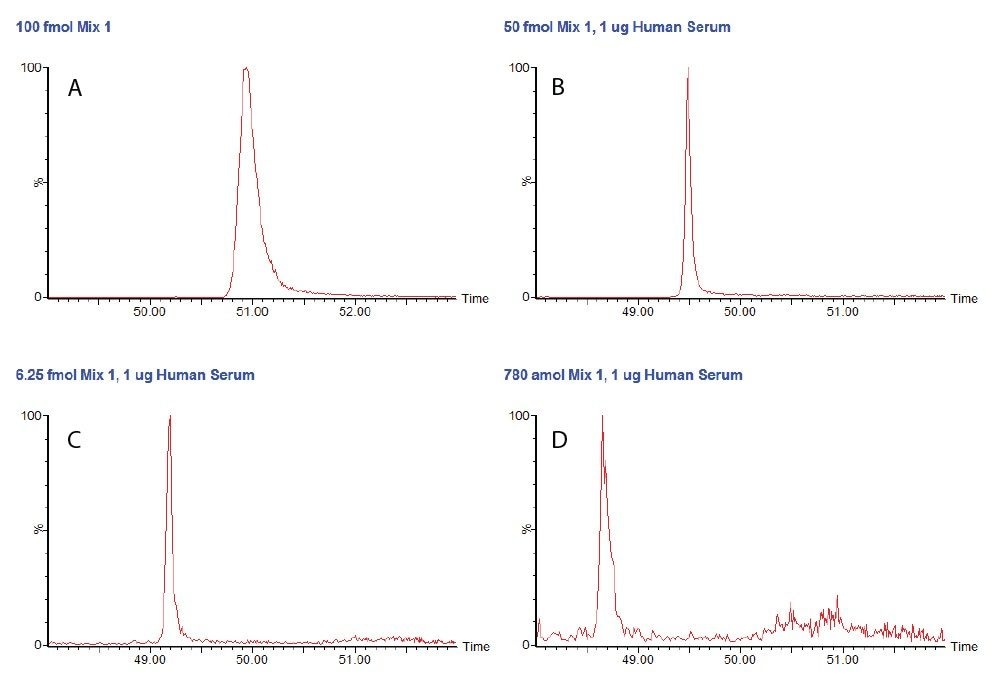
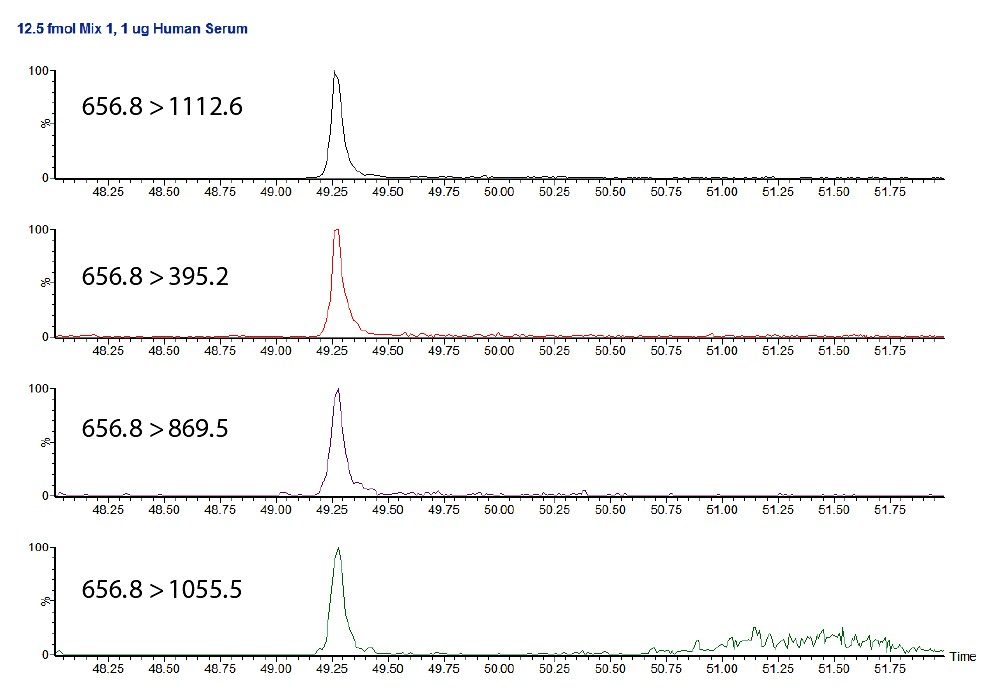
Figure 2 shows the individual transitions for this peptide at 12.5 fmol/μL, where the noise is seen for only the 656.8 to 1055.5 transition. As this is the least intense of the four transitions, it can easily be ignored for the purpose of quantification.
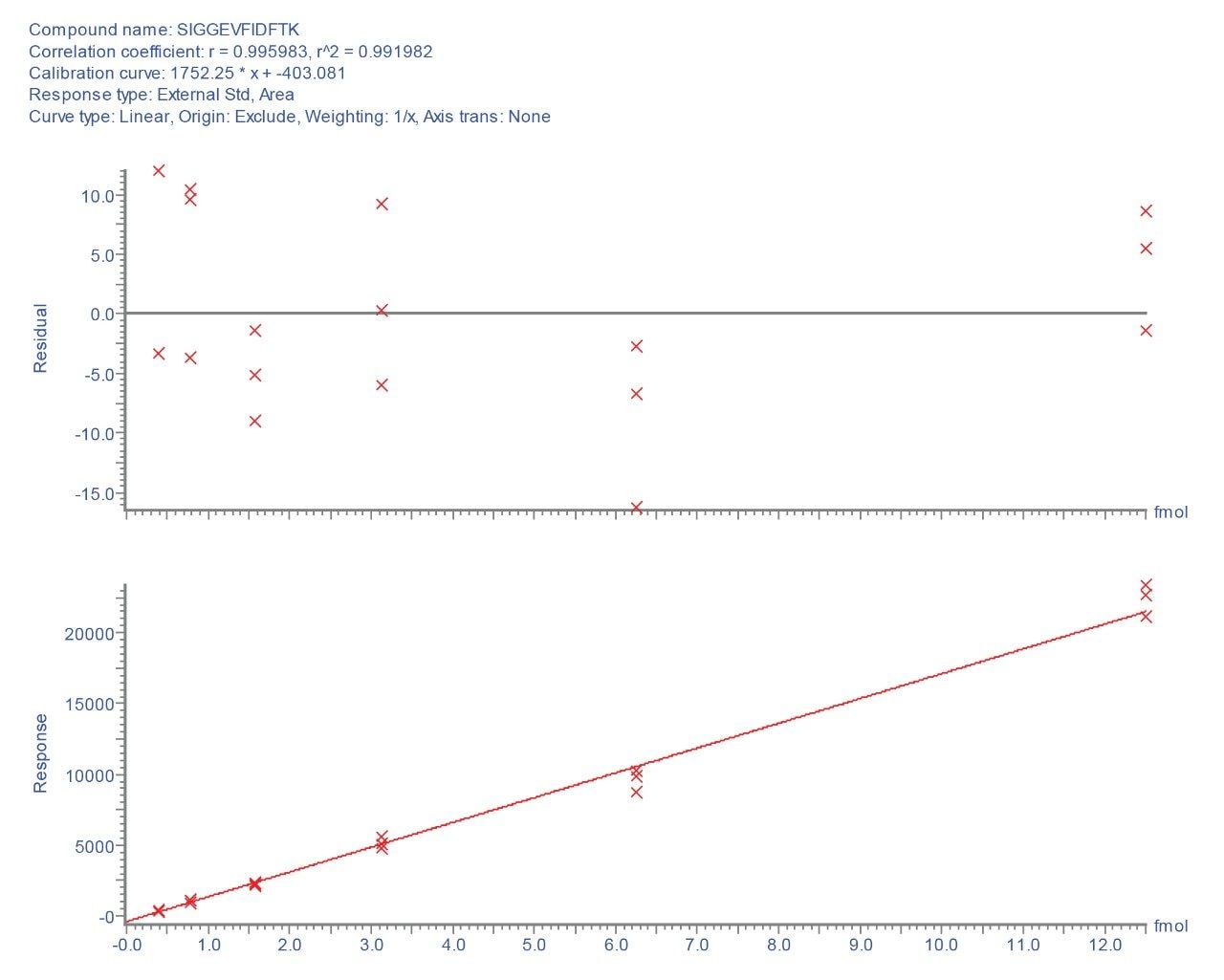
Selecting the most intense transition (656.8 to 1112.6), TargetLynx Application Manager was used to generate a calibration plot for this peptide. Figure 3 shows the resulting standard curve and residuals plot for the critical low concentration range, where a linear fit with r2 > 0.99 is obtained over the range of 390.0 attomoles to 12.5 femtomoles injected.
Method development for quantitative analysis of peptides in complex biological matrices must be rigorous to ensure that the selected peptides and MRM transitions are specific for the protein of interest. The method must also be sufficiently intense to achieve the desired limit of quantitation, and be free of interferences, which could produce erroneous results. When all the criteria are met, the results can be outstanding. The results illustrated here show the case of a peptide for yeast alcohol dehydrogenase, where quantification of less than a femtomole can easily be achieved with 75 μm scale chromatography using a nanoACQUITY UPLC System, coupled with a Xevo TQ MS.
720003883, February 2011