Influenza vaccines provide protection by neutralizing antibodies to viral hemagglutinin (HA), a protein that plays a critical role in influenza infection. In this application note, we demonstrate that UPLC-MSE is capable of separating and characterizing N-linked glycosylations of HA proteins in a recombinant influenza vaccine candidate expressed from insect cell-baculovirus expression vector system (BEVS). Glycopeptides and glycoforms are separated by an ACQUITY UPLC System at the peptide level, and are detected online by a SYNAPT MS System. The UPLC-MSE data are processed by BiopharmaLynx Software to report N-linked glycosylation information.
Influenza is a viral infection of the respiratory tract. It is one of the leading causes of death in the U.S., killing more than 50,000 people per year.1 Influenza vaccination is a primary prophylactic method and the principal strategy for reducing morbidity and mortality due to seasonal influenza. Vaccines provide protection by neutralizing antibodies to viral hemagglutinin (HA), a protein that plays a critical role in influenza infection.
Licensed, inactivated vaccines for seasonal influenza usually contain a preset amount of HAs – a mixture of H1, H3, and B, the corresponding HA proteins of the three most common viruses – influenza A subtypes H1N1 and H3N2, and influenza B. These HA proteins are glycoproteins, with multiple N-linked glycosylation motifs and multiple glycoforms for each glycosylation site. Detailed characterization and monitoring of glycosylation in HAs is important for both vaccine development and production because of their role in determining the function of influenza binding onto host cells and therefore infection.
Currently, the methods for characterization of N-linked glycosylations include released free glycan analysis2-4 and intact mass analysis.5-7 These methods are useful for analyzing glycoproteins with defined glycosylation sites, such as monoclonal antibodies. Glycan profiling can be performed at the intact protein level (intact mass analysis) or as carbohydrates (free glycan profiling). The glycosylation site can usually be determined by peptide mapping8 after the glycan moieties are enzymatically removed (mostly by PNGase F) because the mass of asparagine (N) residues with glycosylation increase by 1 Da upon deglycosylation.
However, it is a challenge for these methods to characterize glycoproteins with multiple glycosylation sites, such as HAs, because it is difficult to distinguish between glycan moieties that link to different sites in a protein. Furthermore, complex samples like vaccines have multiple N sites with an -NXS/T- motif. The determination of N-linked glycosylation sites by peptide mapping may be extremely challenging because modifications of N sites with a 1 Da mass increase could be due to either glycosylation or deamidation.
Using improved resolution of UPLC, four major N-linked glycoforms of a tryptic peptide prepared from a monoclonal mouse IgG1 antibody tryptic digestion were analyzed by LC/UV-MS.9 The experiment demonstrated that glycosylations could be detected and quantified. Both glycosylated site and glycan moiety could be characterized simultaneously.
In previous studies,10-11 we demonstrated that tryptic peptide mapping with UPLC-MSE was capable of separating and characterizing site-specific modifications such as N-deamidation and M-oxidation in an unbiased manner.
In this application note, we demonstrate that UPLC-MSE is capable of separating and characterizing N-linked glycosylations of HA proteins in a recombinant influenza vaccine candidate expressed from insect cell-baculovirus expression vector system (BEVS). Glycopeptides and glycoforms are separated by an ACQUITY UPLC System at the peptide level, and are detected online by a SYNAPT MS System. The UPLC-MSE data are processed by BiopharmaLynx Software to report N-linked glycosylation information. The method offers a way to improve the characterization as well as reduce the amount of time spent on data processing. Furthermore, having a general technique that can be applied to such problems opens this type of analysis to non-experts and could benefit an organization by streamlining their work.
A tryptic digest was prepared for the influenza vaccine candidate sample containing HA proteins H1, H3, and B that were expressed from insect cell-BEVS system. The peptide mixture contained N-linked glycopeptides together with other peptides from the proteins. The preparation procedure included:
The digest was diluted to 0.2 μg/μL with 0.1% FA in 5% acetonitrile (ACN) for UPLC-MSE analysis.
UPLC-MSE experiments were performed using a SYNAPT MS System coupled with an ACQUITY UPLC System. The UPLC system was configured with a 2.1 x 150 mm, BEH300 C18 1.7-μm Peptide Separation Technology column. About 4 μg of the peptide mixture in a 20-μL volume was injected, and eluted using a 120-min gradient (1 to 40% ACN in 0.1% FA) at a flow rate of 0.2 mL/min and column temperature of 60 °C. Four injections were repeated.
MSE data were acquired in ESI positive ion mode, with collision cell energy alternating between low energy (5 V) to collect peptide precursor (MS) data and elevated energy (ramping from 20 to 40 V) to obtain peptide fragmentation (MSE) data. The scan time was 0.5 sec (1 sec total duty cycle). Capillary voltage of 3.0 kV, source temperature of 100 °C, cone voltage of 37 V, and cone gas flow of 10 L/h were maintained during the analyses. The system was tuned for a minimum resolution of 10,000 (V-mode) and calibrated using a 100 fmol/μL Glu1-fibrinopeptide B (GFP) infusion. Sampling of the lock spray channel (100 fmol/μL GFP in 50:50 ACN/water containing 0.1% FA) was performed every 1 min to ensure high mass accuracy.
The collected data were processed by BiopharmaLynx, v.1.2, an application manager for MassLynx Software, using a strict tryptic cleavage rule, and setting cysteine carbamidomethylation as a fixed modification and N-linked glycosylations as variable modifications. Additional BiopharmaLynx method settings were detailed in a previous publication.12
It has been reported13 that glycoproteins expressed from insect cell lines present two major types of N-linked glycans: paucimannosidic structures (Man(1-3)GlcNAc2 or Man(1-3)GlcNAc[Fuc]GlcNAc) and oligomannose structures (Man(5-9)GlcNAc2), where Man represents mannose, GlcNAc is N-acetylglucosamine, and Fuc is fucose. All 11 possible glycosylation forms were entered as variable N-linked glycosylation modifications for BiopharmaLynx data processing. The number of N sites available for N-linked glycosylation (with -NXS/Tmotif) are 9, 12, and 10 in HA proteins H1, H3, and B, respectively. After BiopharmaLynx processing of the collected UPLC-MSE data, we were able to identify 7, 4, and 6 N sites with glycosylation for H1, H3, and B, respectively. Each N-linked glycosylation site may have multiple attached glycoforms. The following are three typical examples (one from each HA protein in the vaccine sample) selected to demonstrate how UPLC-MSE can separate and characterize glycopeptides and glycoforms.
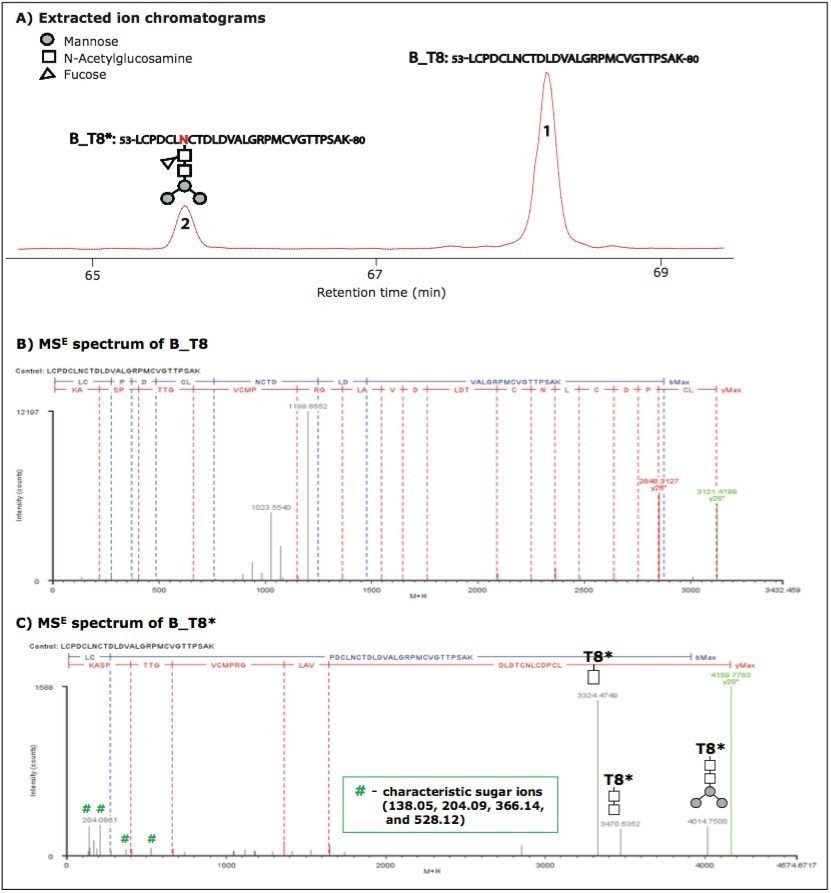
Figure 1 shows the separation and identification of unmodified and glycosylated tryptic peptide T8 in HA protein B (named B_T8 and B_T8*, respectively; in the following text, all glycosylated peptides are marked with an asterisk (*) to signify a corresponding modified form).
B_T8* eluted earlier than B_T8 because the attached sugar group increases hydrophilicity (Figure 1A). The tryptic peptide sequence was confirmed by MSE spectra (Figures 1B and 1C). The glycosylation of B_T8* was indicated by a series of characteristic sugar ions at m/z 138.05, m/z 204.09, m/z 366.14, and m/z 528.12 in the low m/z range and confirmed by ion y28. The 1038.36 mass difference of y28 before and after glycosylation corresponds to a glycan moiety (-Man3NAcGlc[Fuc]NAcGlc). The fragment ions with sugar groups at high m/z range give further structural information about the attached glycan moiety. Based on the MS signal intensities processed by BiopharmaLynx and extracted ion chromatographic areas of precursor masses, the relative concentration of B_T8* is 20%.
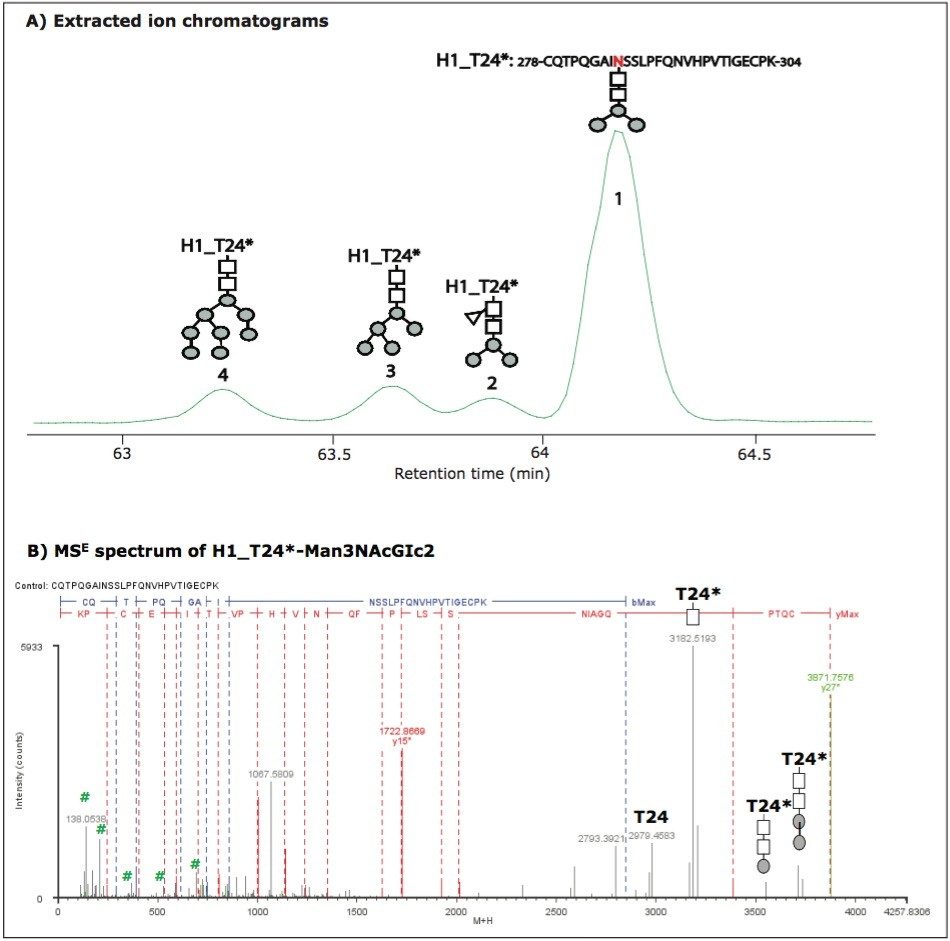
Unlike B_T8*, most identified glycopeptides were fully glycosylated with multiple glycoforms. For example, peptide T24 of HA protein H1. No unmodified H1_T24 was identified, but four glycoforms of H1_T24* were chromatographically resolved and identified (Figure 2A). The glycoform elution order correlated to the size of glycan moiety. The heavier the glycan moiety, the earlier the glycoform eluted for this glycosylated peptide. The peptide has two N sites. MSE spectra confirmed that the glycosylation only occurred on N286 which has the -NSS- motif, and that no glycans attached to N293 with the -NVH- motif. This is consistent with the rule that N-linked glycosylations only occur on N sites with -NXS/T- motif. The MSE spectrum of H1_T24*-Man3NAcGlc2 is shown in Figure 2B. Again, the characteristic sugar ions in the low m/z range and the fragment ions with sugar groups in the high m/z range further confirm the glycosylation and provide structural information of the glycan moiety.
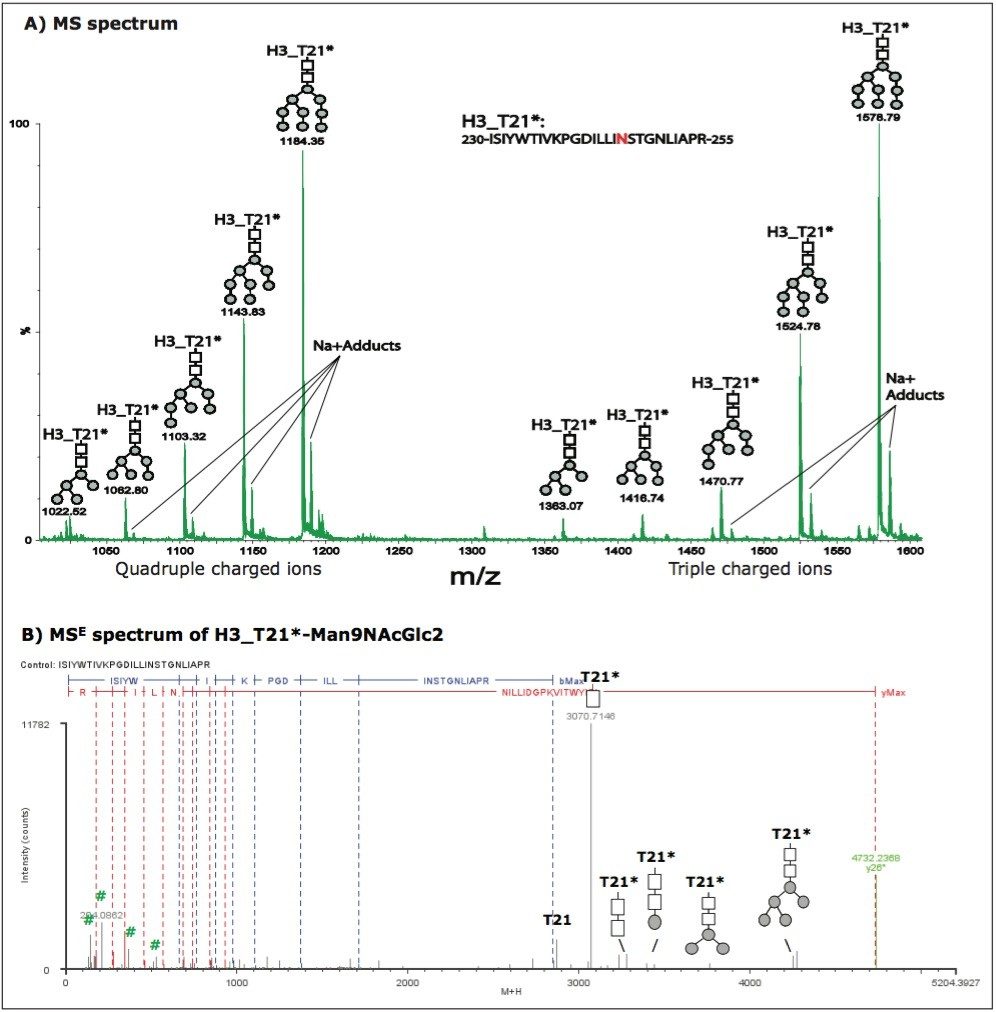
However, not all identified glycoforms were chromatographically resolved. For example, five glycoforms were identified on tryptic peptide T21 from HA protein H3 (see Figure 3A), but these glycoforms were eluted in one LC peak at 91.65 min. Their retention times show small differences (~ 2 seconds) in the extracted ion chromatograms (data not shown). This evidence and the example described above indicate that the chromatographic behavior of glycoforms is related to the nature of the peptide sequence to which the glycan is attached. The peptide sequence of H3_T21 and glycosylations could be confirmed by MSE spectra. An example MSE spectrum of H3_T21*-Man9NAcGlc2 was plotted in Figure 3B.
The results presented here demonstrate that UPLC-MSE can separate and characterize multiple glycopeptides and multiple glycoforms on hemagglutinin glycoproteins in an influenza vaccine sample. Because HA proteins have multiple glycosylation sites and multiple glycoforms, characterization of glycosylations in influenza vaccine samples would be a challenging task for traditional methods such as glycan analysis and intact mass analysis.
Unlike traditional glycosylation characterization methods, in this work the glycosylation sites were unambiguously identified utilizing the MSE technique. MSE also provided useful structural information of glycan moieties from the glycan fragments. In addition, the methods do not involve any additional enrichment or purification procedure, and the data acquired can be automatically processed by BiopharmaLynx.
Further optimization could make this method applicable not only to the characterization of glycosylations in complex samples such as an influenza vaccine, but also as a rapid and routine method for characterizing glycoproteins.
720003173, August 2009