
This is an Application Brief and does not contain a detailed Experimental section.
This application brief highlights the capabilities of automated peak identification, library searching, and chromatogram comparison software within complex mixture analysis.
Rapid detection, identification, and semi-quantitative determination of all components in complex mixtures
Complex mixture analysis is a term that is applicable to a wide-range of MS application areas. A prerequisite is efficient chromatographic separation, whether performed by GC or ACQUITY UPLC. Typically data is acquired in full scan mode on a quadrupole or time-of-flight (ToF) mass spectrometer, such as the Waters GCT Premier or LCT Premier XE. ToF offers significant benefits such as improved full scan sensitivity, reduced cycle time, and high resolution.
The primary challenges for an analyst when reviewing acquired data are:
Each of these processes is time-consuming when performed manually, often resulting in a large number of printed chromatograms, mass spectra, library search results, and compound lists. A single data file could take hours to process, having only taken a few minutes to acquire, with a high probability for error during the manual process.
This technical note shows examples of the use of ChromaLynx XS Software for complex mixture analyses including:
ChromaLynx XS offers a number of automated features to reduce the amount of time taken for these processes, and minimizes the possibility for errors compared with manual processing. Primary features include:
Some representative data from the GCT Premier and LCT Premier XE were used, along with data acquired in EI+ and ESI+/- ionization modes.
All data were acquired using Waters MassLynxTM Software v. 4.1, with data processed using the ChromaLynx XS Application Manager.
Within ChromaLynx XS, acquired raw data files are processed from the sample list user interface, generating a single browser file that contains all of the information about the deconvoluted results:
Figure 1 presents typical complex GC-MS and LC-MS chromatograms, which can be seen to contain a large number of eluting peaks. To manually process these samples, identifying all 100 plus major peaks would take a considerable amount of time.
This process would require the generation of clean background subtracted spectra, sending the spectrum to a library search engine, and then collating the resultant library results. This does not include the added difficulties associated with deconvoluting close or partially co-eluting peaks or having to return to a previously searched peak. Automation of this process can save both time and reduce errors.

Figure 2 shows the ChromaLynx XS browser window with identified peaks denoted using colored pointers. For the complex GC-MS chromatogram shown in Figure 1, ChromaLynx XS has automatically located, library searched, and exact mass scored a few hundred peaks in a matter of minutes.
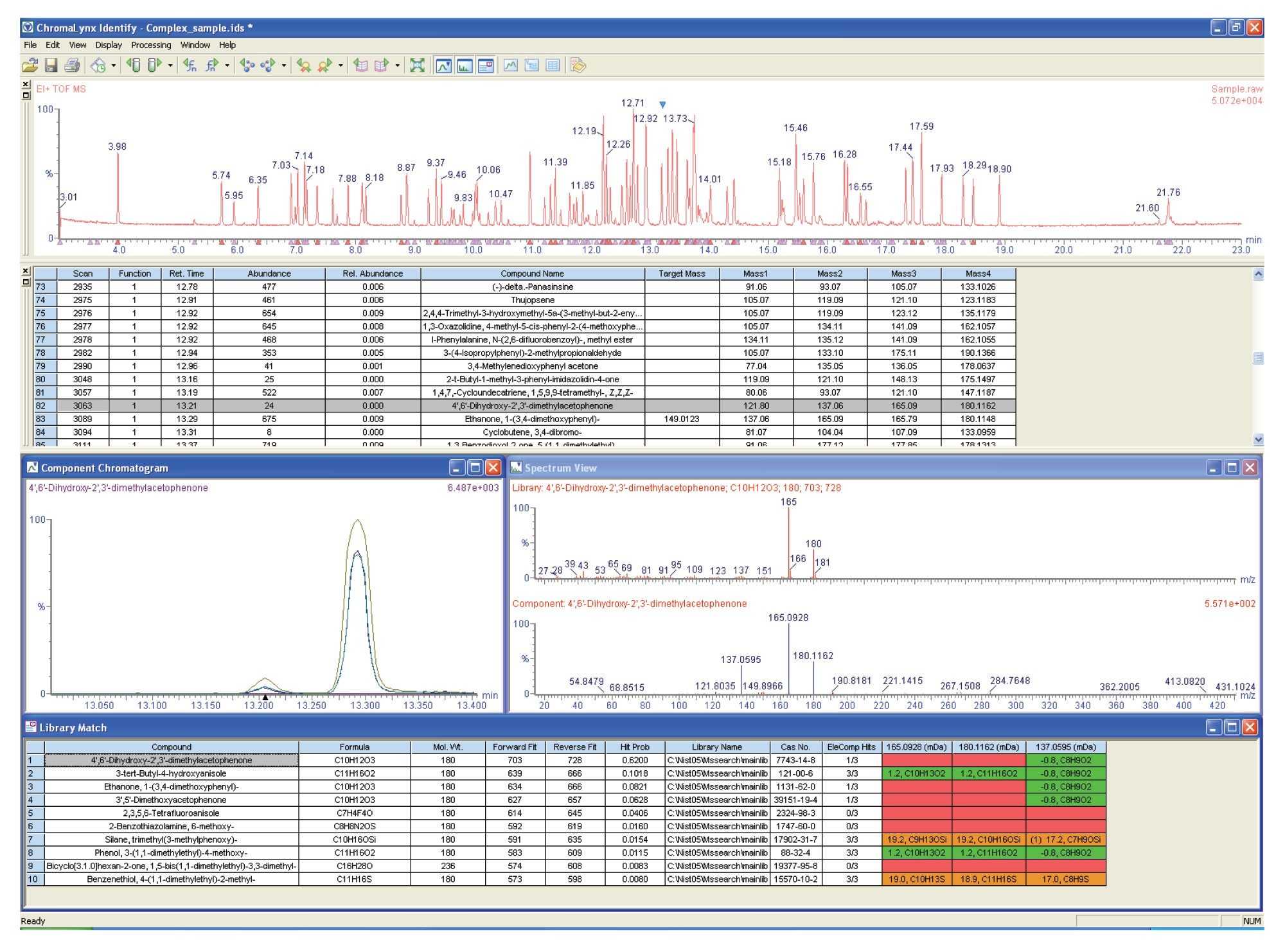
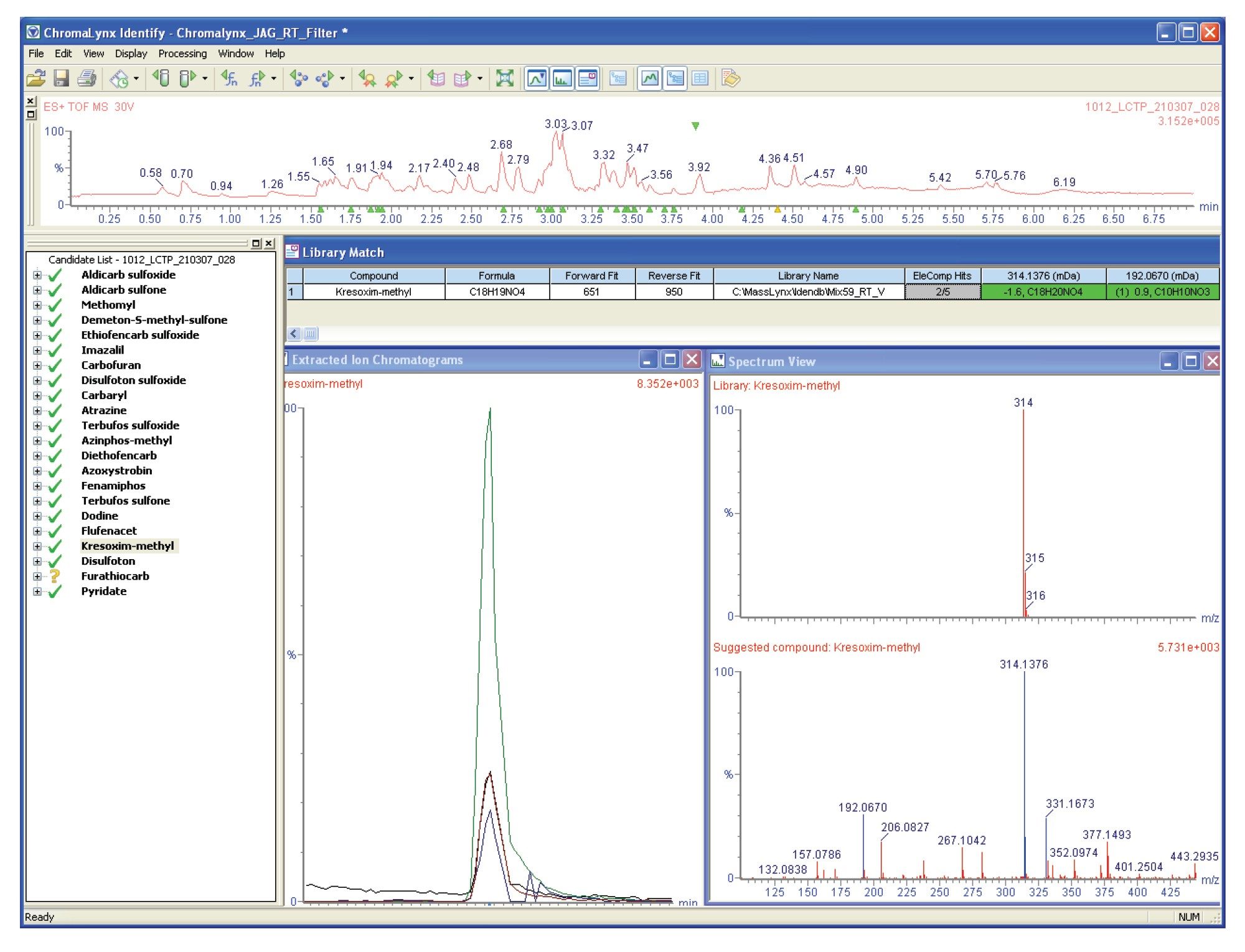
Figure 3 shows the ChromaLynx browser window for the complex UPLC-MS chromatogram, using an alternative candidate screening display, where only the compounds within a library are identified and highlighted.
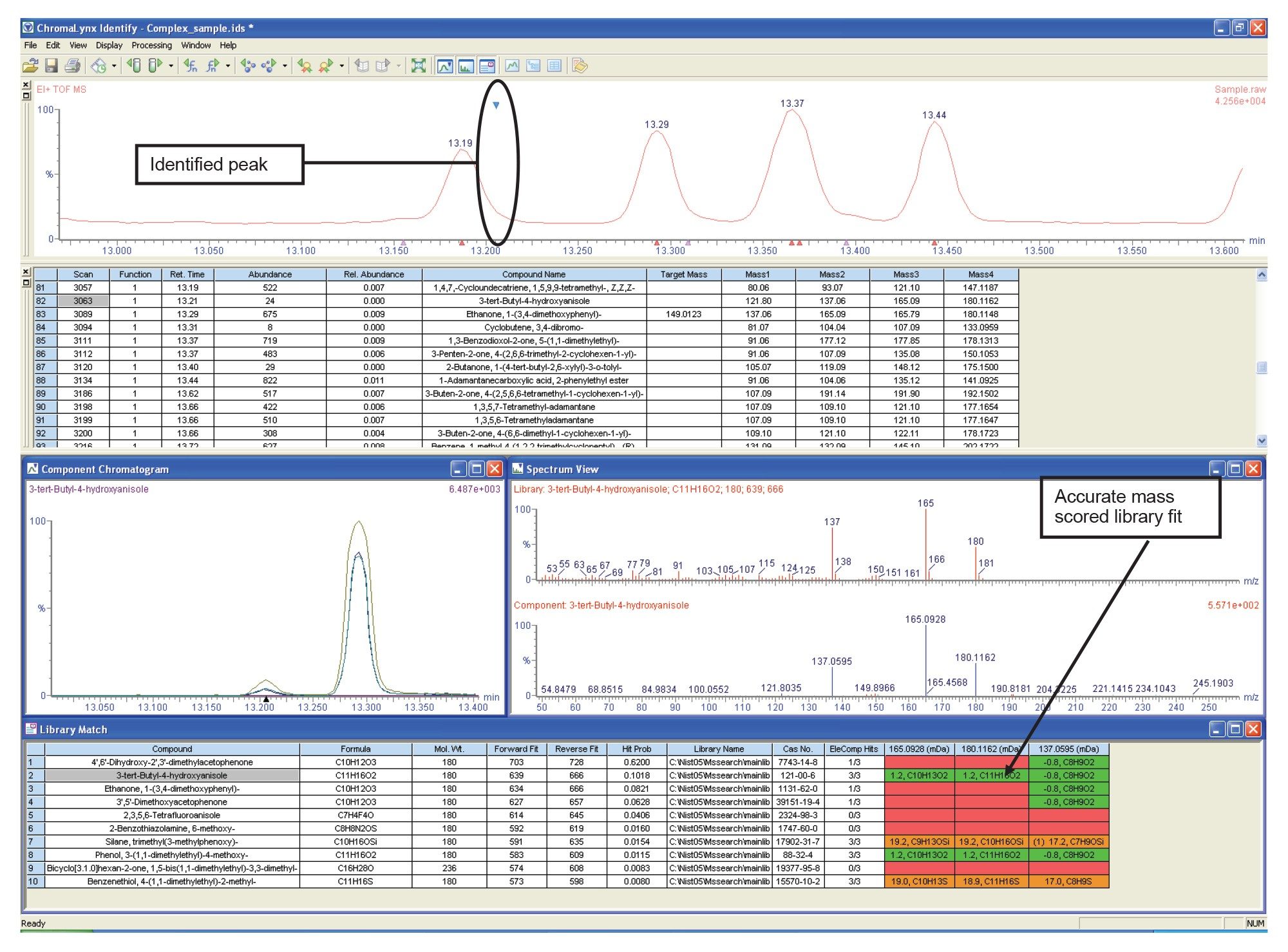
Automated library fit results can be scored according to the exact masses acquired using high resolution ToF instruments. The correct library fit is difficult to assign using nominal mass information only. As shown in figure 4 (highlighted in green), nine out of the ten library fits have a molecular mass of 180, which correspond to the molecular ion in the acquired mass spectrum.
When comparing the acquired mass spectrum with the library spectra, it is difficult to assign a library fit with a high degree of certainty. This case highlights a situation where the high fullspectrum sensitivity of ToF has allowed a very low intensity peak to be detected. Because of the low intensity, it is very difficult to obtain good library fit results.
If the data had been acquired with nominal mass information only, selection of a tentative library fit would not be easy. By applying the exact mass capability, it is possible to propose that the most likely library hits are the result of compounds having the elemental composition C11H16O2 which can now be easily distinguished from the compounds proposed by library search alone.
Accurate mass scoring eases this process by automatically submitting each library entry’s molecular composition to elemental composition calculation software. Within the processing setup, two thresholds can be specified:
1. The first threshold determines the mass accuracy that would give tentative agreement between the acquired and theoretical masses (low mass error).
2. A second threshold that specifies the mass deviation above which an acquired mass is high.
The deconvoluted masses for the acquired spectrum are then displayed, highlighted using colored backgrounds. Green shows a deviation of between zero and the tentative (low) threshold; amber shows a deviation between the low and high thresholds; and red shows where the mass deviation is above the high threshold. In this case, the second and eighth library fits are supported by the exact mass scoring of the molecular and fragment ions, as shown in Figure 4. Manually generating this information would be laborious, with a high probability of error.
Manual comparison of chromatograms is another time-consuming process that can be automated by using ChromaLynx XS. When using the Compare feature, reports can be generated that specify what the common or unique components in complex mixtures are. This is a common process when investigating complaints within the flavor and fragrance, food, fine chemicals, or environmental industries.
Often the differences between chromatograms can highlight issues resulting from adulteration, tainting, or contamination of products or sample matrices (essential oils, soil, drinking water, etc.).
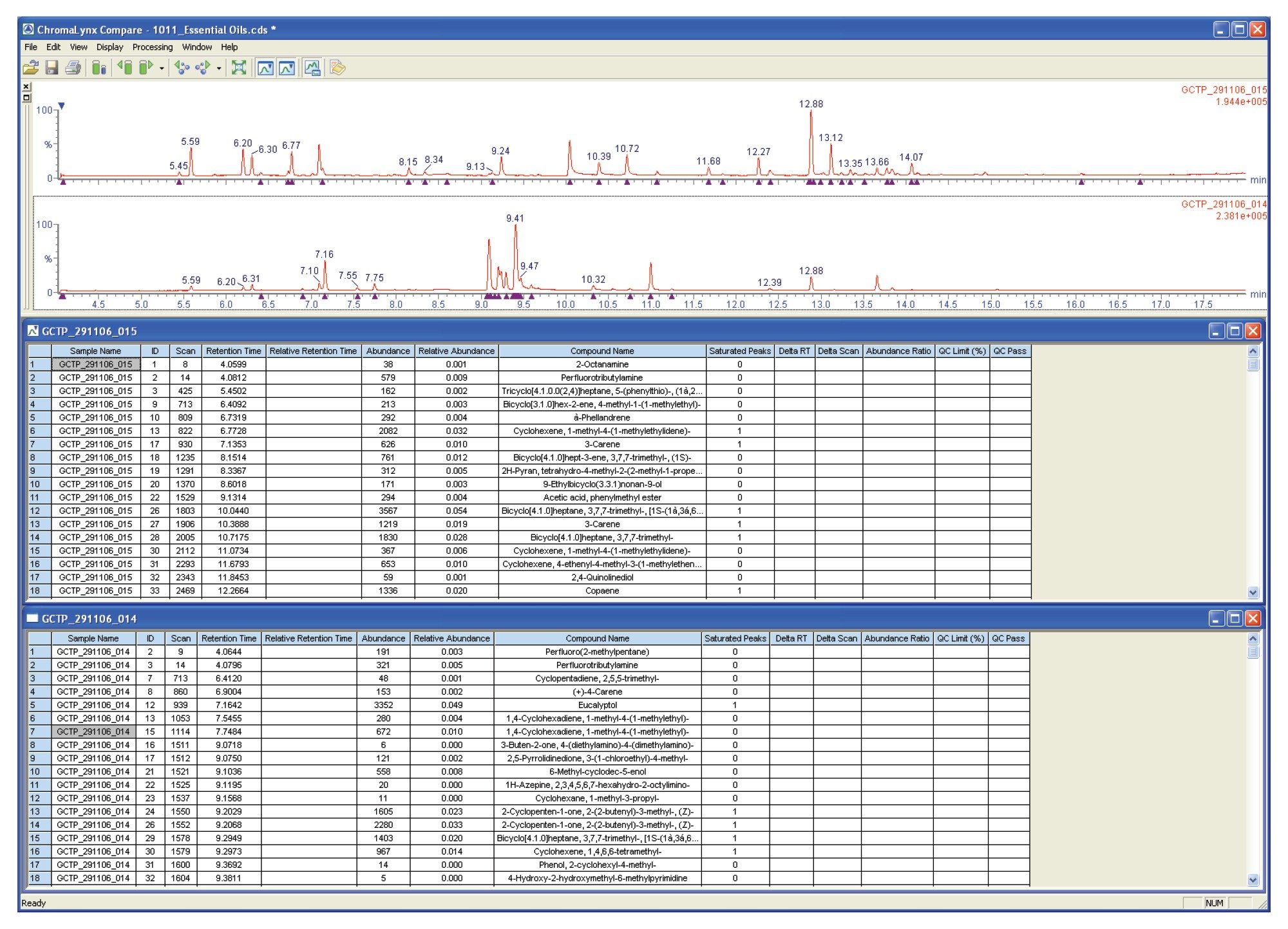
The browser window shown in Figure 5 compares a premix essential oil with a peppermint essential oil. Although ChromaLynx XS does not perform detailed quantification, it can compare peak areas, either against each other or against the total ion count (TIC). Here, the two complex mixtures have been compared on a mass and retention time basis, with QC scoring highlighting a pair of common peaks that also have similar area counts (in this case, a difference of less than 20% between the two samples).
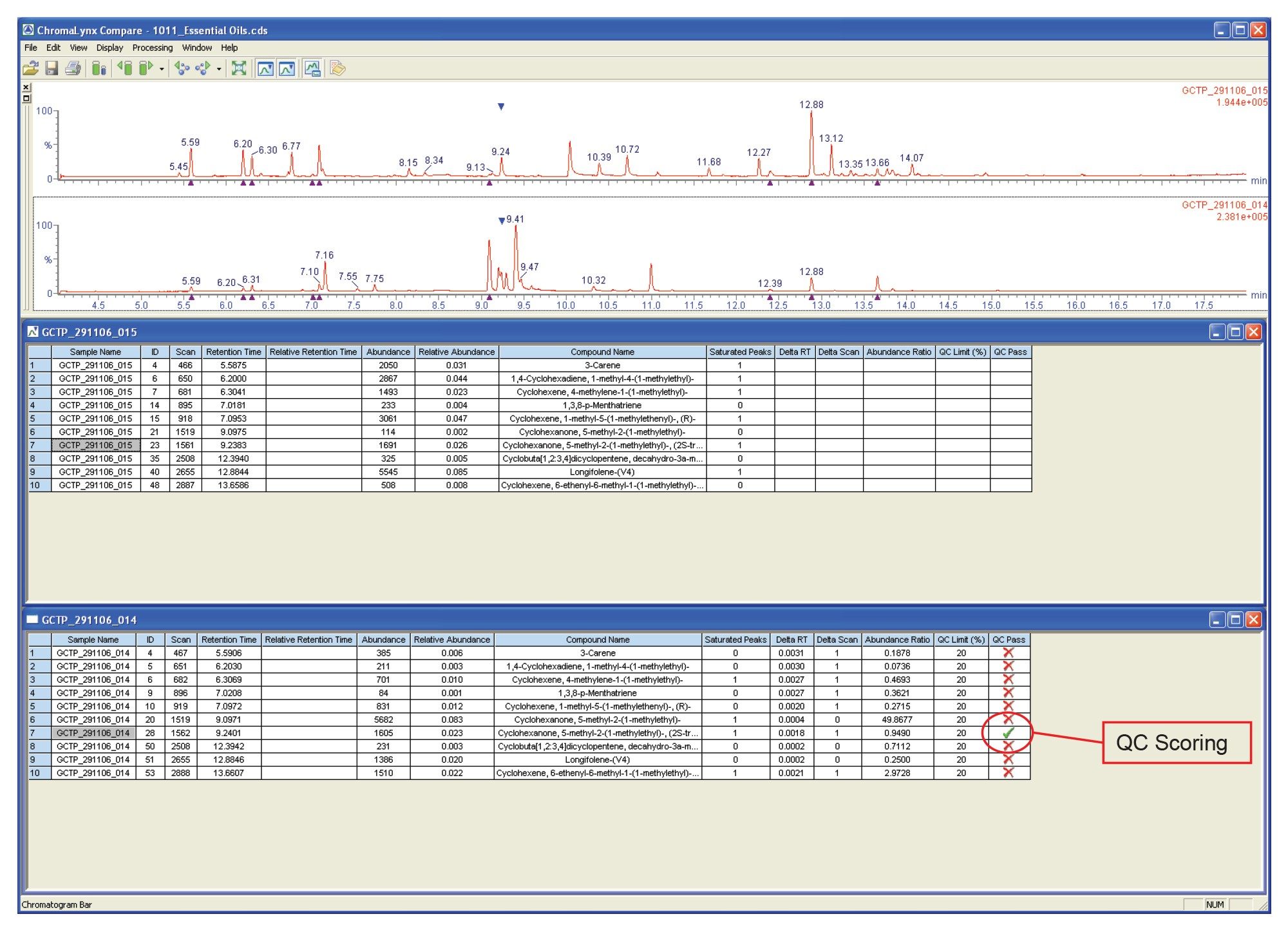
This QC scoring is highlighted by assigning a green tick to the peak in question. The Compare report shows that there are not many common peaks, indicating that ten compounds are common with one compound present at a similar intensity when comparing the premix and peppermint essential oils.
ChromaLynx XS Compare can also display the unique components detected within different samples, as shown in Figure 6. In this case, peppermint oil is not one of the constituent components of the premix oil, so a much larger number of unique components are being highlighted.
- The rapid detection, identification, and semi-quantitative determination of all components in complex mixtures.
- The combination of non-targeted component detection with a library search to facilitate identification.
720002643, July 2008